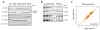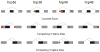Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members - PubMed (original) (raw)
Genome-wide analysis of alternative pre-mRNA splicing and RNA-binding specificities of the Drosophila hnRNP A/B family members
Marco Blanchette et al. Mol Cell. 2009.
Abstract
Heterogeneous nuclear ribonucleoproteins (hnRNPs) have been traditionally seen as proteins packaging RNA nonspecifically into ribonucleoprotein particles (RNPs), but evidence suggests specific cellular functions on discrete target pre-mRNAs. Here we report genome-wide analysis of alternative splicing patterns regulated by four Drosophila homologs of the mammalian hnRNP A/B family (hrp36, hrp38, hrp40, and hrp48). Analysis of the global RNA-binding distributions of each protein revealed both small and extensively bound regions on target transcripts. A significant subset of RNAs were bound and regulated by more than one hnRNP protein, revealing a combinatorial network of interactions. In vitro RNA-binding site selection experiments (SELEX) identified distinct binding motif specificities for each protein, which were overrepresented in their respective regulated and bound transcripts. These results indicate that individual heterogeneous ribonucleoproteins have specific affinities for overlapping, but distinct, populations of target pre-mRNAs controlling their patterns of RNA processing.
Figures
Figure 1
Knockdown of expression of four members of the Drosophila hnRNP A/B family by RNAi in Drosophila S2 cells. A) Western blot analysis shows that hrp36, hrp38, hrp40 and hrp48 are efficiently knock down in triplicate samples compare to cells treated with a non-specific dsRNA (NS). Alpha-tubulin is used as control to confirm that similar levels of proteins were loaded. B) Titration of the protein sample treated with non-specific dsRNA is used to evaluate the knockdown efficiency. C) The overall changes in the net-expression value is similar for cells knockdown in the hrp36 expression with two different non-overlapping dsRNA. A splicing junction in one knockdown is plotted against the same splicing junction in the second knock down. The relative feature density is expressed as darker shade of yellow. The blue line is the fitted linear model used to calculate the R-value. The dashed red line corresponds to a perfect correlation.
Figure 2
Genome-wide identification of splice junctions regulated by individual members of the Drosophila hnRNP A/B family. A) Venn diagram of the extent of overlap of the splice junctions regulated by each individual hnRNP protein. B) Representation of the co-regulated junctions for each splicing regulator demonstrating that most of the co-regulated junctions are affected in the same direction (green) or the opposite direction (red) together with the fraction of the junctions co-regulated for each individual hnRNP protein. C) Breakdown of the number of junctions found to be significantly affected in each individual RNAi knockdown. Constitutive junctions are shown in light blue (up) and dark blue (down). Alternative junctions are shown in light orange (up) or red (down). D) Number of genes with junctions regulated by a single hnRNP protein (specific-light yellow) together with the fraction of genes co-regulated by a second hnRNP protein (shared-red).
Figure 3
Genome-wide identification of the RNAs associated with the different hnRNP proteins in RNP complexes. A) Distribution of the level of gene expression measured by Affymetrix arrays measured in S2 cells (blue bar) and the fraction of RNA bound by a hnRNP protein within a given level of expression (purple bar). B) The overlap between the genes that were found regulated in the splice junction microarray (blue) and bound in the tiling array (yellow). The highly significant overlapping transcripts are represented in brown (Fisher exact test, p-value<2×10−5). D) TepII and pes, two representative genes bound by hnRNP proteins showing extensive coverage suggestive of the spreading model. C) Fur1 a representative gene bound by hnRNP proteins showing discrete intronic binding tracts for hrp40 suggestive of the looping model. Significantly over-represented signal in the IP samples over the signal measured from RNA extracted from the starting RNP extract are represented by bars along the genome with the eight of the bar corresponding to p-values (expressed as −Log (p)). Above is a representation of the gene structure, blue boxes are constitutive exons, red boxes are alternative exons, lines are introns. Pes has two alternative promoters while Fur1 as two alternative poly-A signals represented by the 2 levels in the gene structure.
Figure 4
HnRNP proteins bind to different and specific RNA motifs that are over-represented in the genes regulated and bound by the respective hnRNP proteins. A) Graphical representation of the preferred binding motif identified by SELEX for each individual hnRNP protein (Crooks et al., 2004). Height of each bar shows the information content at each position of the binding motif in bits (log-odds in base 2). B) Cumulative distribution of the presence of at least three occurrences of the SELEX motif in the genes identified as being regulated by the splice junction array or identified as bound by the tiling array. Error bars are determined analytically using the binomial distribution and correspond to 1 standard deviation: sqrt[np(1 − p)]/n, where n is the number of sequences searched and p is the observed probability of having sites of given score. C–D) Hph and psq, two representative genes showing that the binding tracts identified on the tiling arrays (colored bar tracts) are frequently associated with clusters of high affinity binding sites. A moving window using the SELEX motifs was used to calculate the score at each position along the fly genome. The motif score at a given position is represented as a heat map, blue low motif score-yellow high motif score. E) Magnification of Hph binding tracts showing a 20,000 nt long region (middle panel) to the nucleotide level showing the underlying genomic sequence (bottom of the window) with the presence of the SELEX motifs (colored bar above the sequence) corresponding to the hrp48 and hrp40 recognized motifs (cyan and magenta boxes respectively).
Figure 4
HnRNP proteins bind to different and specific RNA motifs that are over-represented in the genes regulated and bound by the respective hnRNP proteins. A) Graphical representation of the preferred binding motif identified by SELEX for each individual hnRNP protein (Crooks et al., 2004). Height of each bar shows the information content at each position of the binding motif in bits (log-odds in base 2). B) Cumulative distribution of the presence of at least three occurrences of the SELEX motif in the genes identified as being regulated by the splice junction array or identified as bound by the tiling array. Error bars are determined analytically using the binomial distribution and correspond to 1 standard deviation: sqrt[np(1 − p)]/n, where n is the number of sequences searched and p is the observed probability of having sites of given score. C–D) Hph and psq, two representative genes showing that the binding tracts identified on the tiling arrays (colored bar tracts) are frequently associated with clusters of high affinity binding sites. A moving window using the SELEX motifs was used to calculate the score at each position along the fly genome. The motif score at a given position is represented as a heat map, blue low motif score-yellow high motif score. E) Magnification of Hph binding tracts showing a 20,000 nt long region (middle panel) to the nucleotide level showing the underlying genomic sequence (bottom of the window) with the presence of the SELEX motifs (colored bar above the sequence) corresponding to the hrp48 and hrp40 recognized motifs (cyan and magenta boxes respectively).
Figure 5
Locations of binding site enrichment for each hnRNP. Circles show positions that had an enrichment of SELEX motifs in affected splicing events (two-sided Wilcoxon ranked-sum test, p-value<0.01 without multiple trials correction for >2,000 trials) using any of several measures for enrichment (see Experimental Procedures and Supp. Figure 8 for details).
Figure 6
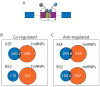
No significant antagonistic regulation is found between two SR proteins and four hnRNP A/B family proteins in a single defined cellular environment. A) It has been suggested that several genes are under the antagonistic control of SR and hnRNP proteins where the pattern of splice isoforms is dependent on the relative concentrations of SR and hnRNP proteins. B) Comparison of the genes that are found to be regulated in the same direction between hnRNPs (this analysis) and two different SR proteins (Blanchette et al., 2005). B) Comparison of the genes that are found to be regulated in opposite directions between hnRNPs (this analysis) and two different SR proteins (Blanchette et al., 2005).
Similar articles
- Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo.
Zu K, Sikes ML, Haynes SR, Beyer AL. Zu K, et al. Mol Biol Cell. 1996 Jul;7(7):1059-73. doi: 10.1091/mbc.7.7.1059. Mol Biol Cell. 1996. PMID: 8862520 Free PMC article. - Genome-Wide Analysis of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) Binding to HIV-1 RNA Reveals a Key Role for hnRNP H1 in Alternative Viral mRNA Splicing.
Kutluay SB, Emery A, Penumutchu SR, Townsend D, Tenneti K, Madison MK, Stukenbroeker AM, Powell C, Jannain D, Tolbert BS, Swanstrom RI, Bieniasz PD. Kutluay SB, et al. J Virol. 2019 Oct 15;93(21):e01048-19. doi: 10.1128/JVI.01048-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31413137 Free PMC article. - An intron element modulating 5' splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1.
Chabot B, Blanchette M, Lapierre I, La Branche H. Chabot B, et al. Mol Cell Biol. 1997 Apr;17(4):1776-86. doi: 10.1128/MCB.17.4.1776. Mol Cell Biol. 1997. PMID: 9121425 Free PMC article. - hnRNP A1: the Swiss army knife of gene expression.
Jean-Philippe J, Paz S, Caputi M. Jean-Philippe J, et al. Int J Mol Sci. 2013 Sep 16;14(9):18999-9024. doi: 10.3390/ijms140918999. Int J Mol Sci. 2013. PMID: 24065100 Free PMC article. Review. - Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles.
Bekenstein U, Soreq H. Bekenstein U, et al. Mol Cell Neurosci. 2013 Sep;56:436-46. doi: 10.1016/j.mcn.2012.12.002. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247072 Review.
Cited by
- RNA processing and its regulation: global insights into biological networks.
Licatalosi DD, Darnell RB. Licatalosi DD, et al. Nat Rev Genet. 2010 Jan;11(1):75-87. doi: 10.1038/nrg2673. Nat Rev Genet. 2010. PMID: 20019688 Free PMC article. Review. - Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches.
Chen M, Manley JL. Chen M, et al. Nat Rev Mol Cell Biol. 2009 Nov;10(11):741-54. doi: 10.1038/nrm2777. Epub 2009 Sep 23. Nat Rev Mol Cell Biol. 2009. PMID: 19773805 Free PMC article. Review. - Identification and experimental validation of splicing regulatory elements in Drosophila melanogaster reveals functionally conserved splicing enhancers in metazoans.
Brooks AN, Aspden JL, Podgornaia AI, Rio DC, Brenner SE. Brooks AN, et al. RNA. 2011 Oct;17(10):1884-94. doi: 10.1261/rna.2696311. Epub 2011 Aug 24. RNA. 2011. PMID: 21865603 Free PMC article. - Evolution of a tissue-specific splicing network.
Taliaferro JM, Alvarez N, Green RE, Blanchette M, Rio DC. Taliaferro JM, et al. Genes Dev. 2011 Mar 15;25(6):608-20. doi: 10.1101/gad.2009011. Genes Dev. 2011. PMID: 21406555 Free PMC article. - Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.
Roesmann F, Müller L, Klaassen K, Heß S, Widera M. Roesmann F, et al. Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review.
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. - PubMed
- Aznarez I, Barash Y, Shai O, He D, Zielenski J, Tsui LC, Parkinson J, Frey BJ, Rommens JM, Blencowe BJ. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 2008;18:1247–1258. - PMC - PubMed
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
- Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM61987/GM/NIGMS NIH HHS/United States
- R01 GM071655/GM/NIGMS NIH HHS/United States
- R01GM071655/GM/NIGMS NIH HHS/United States
- R01 GM061987/GM/NIGMS NIH HHS/United States
- R01 GM061987-17/GM/NIGMS NIH HHS/United States
- U01HG004271/HG/NHGRI NIH HHS/United States
- U01 HG004271/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous