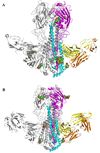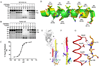Antibody recognition of a highly conserved influenza virus epitope - PubMed (original) (raw)
Antibody recognition of a highly conserved influenza virus epitope
Damian C Ekiert et al. Science. 2009.
Abstract
Influenza virus presents an important and persistent threat to public health worldwide, and current vaccines provide immunity to viral isolates similar to the vaccine strain. High-affinity antibodies against a conserved epitope could provide immunity to the diverse influenza subtypes and protection against future pandemic viruses. Cocrystal structures were determined at 2.2 and 2.7 angstrom resolutions for broadly neutralizing human antibody CR6261 Fab in complexes with the major surface antigen (hemagglutinin, HA) from viruses responsible for the 1918 H1N1 influenza pandemic and a recent lethal case of H5N1 avian influenza. In contrast to other structurally characterized influenza antibodies, CR6261 recognizes a highly conserved helical region in the membrane-proximal stem of HA1 and HA2. The antibody neutralizes the virus by blocking conformational rearrangements associated with membrane fusion. The CR6261 epitope identified here should accelerate the design and implementation of improved vaccines that can elicit CR6261-like antibodies, as well as antibody-based therapies for the treatment of influenza.
Figures
Figure 1. Crystal structures of broadly neutralizing antibody CRF6261 in complex with SC1918/H1 and Viet04/H5 HAs
A) The trimeric CRF6261-SC1918/H1 complex. One HA/Fab protomer is colored with HA1 in purple, HA2 in cyan, Fab heavy chain in yellow, and Fab light chain in orange; the other two HA monomers and Fabs in the trimer are colored in grey for clarity. Glycans are depicted as their component atoms (carbon in green, oxygen in red and nitrogen in blue) with their van der Waals’ radii. B) The trimeric CRF6261-Viet04 complex, colored as in A. The HA1 globular heads are located at the top of the figure and the membrane-proximal stem is at the bottom where the Fab is bound.
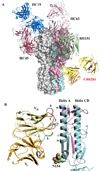
Figure 2. Broadly neutralizing CR6261 binds in the HA stem distant from other strain-specific antibodies using only its heavy chain
A) Comparison of the binding sites of Fab CR6261 (yellow and orange, as in Fig. 1)) that recognizes the HA stem and strain-specific antibodies that bind to the HA1 globular head domain The strain-specific antibody structures are depicted in green (BH151, PDB code 1EO8), copper (HC63, PDB code 1KEN), dark red (HC45, PDB code 1QFU), and blue (HC19, PDB code 2VIR). The HA trimer is shown as a surface representation, with the HA1 and HA2 from one protomer colored in purple and cyan, respectively. B) A close up view of the interaction of CR6261 with the A-helix. HA1 and HA2 are colored in purple and cyan, respectively. The HCDRs 1, 2, and 3 of the CR6261 VH domain are highlighted in red, blue, and green, respectively. In addition, the 72–73 loop from FR3 of VH, which is structurally analogous to the CDR HV4 of a TCR, is indicated in purple. CDR1 runs along the side of the A-helix, interacting with five consecutive helical turns. In contrast, the light chain makes no contacts with the HA and is separated from the nearest HA side chain by ∼8 Å.
Figure 3. Interaction of CR6261 with SC1918/H1 and Viet04/H5 HA illustrating the neutralizing epitope
Surface representations of the interaction of CR6261 with SC1918/H1 (A–C) and Viet04/H5 (D–F). The coloring in A, B, D, and E corresponds to Fig. 1, with interacting epitope residues highlighted in a darker shade. Atoms making polar contacts are highlighted in red. C and F depict the electrostatic potential surface of the epitope on the HA (red – negative; blue-positive; grey-neutral) with HCDRs 1, 2, 3, and FR3 (CDR “HV4”) represented as sticks and colored blue, red, green, and purple, as in Fig. 2. Key interacting residues on the antibody and the HA are indicated on the top of the surface with labels.
Figure 4. CR6261 recognizes a functionally conserved epitope in the stalk region and inhibits the pH-induced conformational changes in the SC1918/H1 and Viet04/H5 HAs
CR6261 protects SC1918/H1 (A) and Viet04/H5 (B) HAs from the pH-induced protease sensitivity associated with membrane fusion. Exposure to low pH renders the SC1918-H1 and Viet04-H5 HAs sensitive to trypsin digestion (lanes 7–8 vs. 9), but CR6261 prevents conversion to the protease susceptible conformation (lanes 10–12). Note that the CR6261-SC1918-H1 crystals were grown at pH 5.3 that also indicates CR6261 blocks the extensive pH-induced conformational changes. C) Titration of SC1918-H1 trypsin-resistance versus varying pH-treatments (followed by neutralization to pH 8.0). The pH resulting in 50% conversion to protease sensitive conformation is 5.76 (95% confidence interval, 5.70–5.82). D) Superposition of the A-helix from SC1918/H1 (green) and an H3 HA (yellow, PDB code 2VIU) reveals that the CR6261 interacting surface is highly conserved among all subtypes. Helix positions are labeled according to the SC1918/H1 sequence, with the percent similarity across all subtypes (H1-16, from an analysis of 5261 sequences) indicated in parentheses. E–F) HA2 undergoes a dramatic and irreversible conformational change between the pre- and post-fusion states. This results in the translocation of the A-helix (red) from its initial position near the viral envelope (Fig. E) towards the target membrane at the opposite end of the HA trimer (Fig. F). The zipping up of coil H along the outside of the A and B-helices is thought to drive the fusion reaction. The orientation of helix C (yellow) is roughly identical in Figs. E and F. G) Polar residues on the CR6261-interacting surface of the A-helix form a network of interactions with coil H in the post-fusion state. These residues are well positioned to play a critical role in the late stages of membrane fusion, explaining the exceptional conservation the CR6261 epitope on the A-helix.
Similar articles
- Unmasking Stem-Specific Neutralizing Epitopes by Abolishing N-Linked Glycosylation Sites of Influenza Virus Hemagglutinin Proteins for Vaccine Design.
Liu WC, Jan JT, Huang YJ, Chen TH, Wu SC. Liu WC, et al. J Virol. 2016 Sep 12;90(19):8496-508. doi: 10.1128/JVI.00880-16. Print 2016 Oct 1. J Virol. 2016. PMID: 27440889 Free PMC article. - Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells.
Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. Throsby M, et al. PLoS One. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942. Epub 2008 Dec 16. PLoS One. 2008. PMID: 19079604 Free PMC article. - A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses.
Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, Calder L, Gamblin SJ, Zhang L, Deubel V, Zhou B, Skehel JJ, Zhou P. Hu H, et al. J Virol. 2012 Mar;86(6):2978-89. doi: 10.1128/JVI.06665-11. Epub 2012 Jan 11. J Virol. 2012. PMID: 22238297 Free PMC article. - Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus.
Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. Xu R, et al. Science. 2010 Apr 16;328(5976):357-60. doi: 10.1126/science.1186430. Epub 2010 Mar 25. Science. 2010. PMID: 20339031 Free PMC article. - The antigenic architecture of the hemagglutinin of influenza H5N1 viruses.
Velkov T, Ong C, Baker MA, Kim H, Li J, Nation RL, Huang JX, Cooper MA, Rockman S. Velkov T, et al. Mol Immunol. 2013 Dec;56(4):705-19. doi: 10.1016/j.molimm.2013.07.010. Epub 2013 Aug 7. Mol Immunol. 2013. PMID: 23933511 Review.
Cited by
- Intranasal antibody gene transfer in mice and ferrets elicits broad protection against pandemic influenza.
Limberis MP, Adam VS, Wong G, Gren J, Kobasa D, Ross TM, Kobinger GP, Tretiakova A, Wilson JM. Limberis MP, et al. Sci Transl Med. 2013 May 29;5(187):187ra72. doi: 10.1126/scitranslmed.3006299. Sci Transl Med. 2013. PMID: 23720583 Free PMC article. - Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines.
Xuan C, Shi Y, Qi J, Zhang W, Xiao H, Gao GF. Xuan C, et al. Protein Cell. 2011 Dec;2(12):997-1005. doi: 10.1007/s13238-011-1134-y. Epub 2012 Jan 10. Protein Cell. 2011. PMID: 22231357 Free PMC article. - Grafting Methionine on 1F1 Ab Increases the Broad-Activity on HA Structural-Conserved Residues of H1, H2, and H3 Influenza a Viruses.
Le HT, Do PC, Le L. Le HT, et al. Evol Bioinform Online. 2021 Mar 16;17:11769343211003082. doi: 10.1177/11769343211003082. eCollection 2021. Evol Bioinform Online. 2021. PMID: 33795930 Free PMC article. - Reconstruction of the 1918 influenza virus: unexpected rewards from the past.
Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG, Wilson IA. Taubenberger JK, et al. mBio. 2012 Sep 11;3(5):e00201-12. doi: 10.1128/mBio.00201-12. Print 2012. mBio. 2012. PMID: 22967978 Free PMC article. Review. - Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses.
Wu NC, Thompson AJ, Lee JM, Su W, Arlian BM, Xie J, Lerner RA, Yen HL, Bloom JD, Wilson IA. Wu NC, et al. Science. 2020 Jun 19;368(6497):1335-1340. doi: 10.1126/science.aaz5143. Science. 2020. PMID: 32554590 Free PMC article.
References
- Schafer JR, et al. Virology. 1993;194:781. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Y1-GM-1104/GM/NIGMS NIH HHS/United States
- Y1-CO-1020/CO/NCI NIH HHS/United States
- P01 AI058113/AI/NIAID NIH HHS/United States
- AI-058113/AI/NIAID NIH HHS/United States
- U54 GM074898/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous