piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells - PubMed (original) (raw)
. 2009 Apr 9;458(7239):766-70.
doi: 10.1038/nature07863. Epub 2009 Mar 1.
Iacovos P Michael, Paria Mohseni, Ridham Desai, Maria Mileikovsky, Riikka Hämäläinen, Rebecca Cowling, Wei Wang, Pentao Liu, Marina Gertsenstein, Keisuke Kaji, Hoon-Ki Sung, Andras Nagy
Affiliations
- PMID: 19252478
- PMCID: PMC3758996
- DOI: 10.1038/nature07863
piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells
Knut Woltjen et al. Nature. 2009.
Abstract
Transgenic expression of just four defined transcription factors (c-Myc, Klf4, Oct4 and Sox2) is sufficient to reprogram somatic cells to a pluripotent state. The resulting induced pluripotent stem (iPS) cells resemble embryonic stem cells in their properties and potential to differentiate into a spectrum of adult cell types. Current reprogramming strategies involve retroviral, lentiviral, adenoviral and plasmid transfection to deliver reprogramming factor transgenes. Although the latter two methods are transient and minimize the potential for insertion mutagenesis, they are currently limited by diminished reprogramming efficiencies. piggyBac (PB) transposition is host-factor independent, and has recently been demonstrated to be functional in various human and mouse cell lines. The PB transposon/transposase system requires only the inverted terminal repeats flanking a transgene and transient expression of the transposase enzyme to catalyse insertion or excision events. Here we demonstrate successful and efficient reprogramming of murine and human embryonic fibroblasts using doxycycline-inducible transcription factors delivered by PB transposition. Stable iPS cells thus generated express characteristic pluripotency markers and succeed in a series of rigorous differentiation assays. By taking advantage of the natural propensity of the PB system for seamless excision, we show that the individual PB insertions can be removed from established iPS cell lines, providing an invaluable tool for discovery. In addition, we have demonstrated the traceless removal of reprogramming factors joined with viral 2A sequences delivered by a single transposon from murine iPS lines. We anticipate that the unique properties of this virus-independent simplification of iPS cell production will accelerate this field further towards full exploration of the reprogramming process and future cell-based therapies.
Figures
Figure 1
Cell lines generated by PB-mediated factor transposition are reprogrammed. (a) The PB-TET transposon vector used to deliver inducible (tet-O), reporter-linked (IRES-ßgeo-pA) mouse factors (mFx). 3′/5′TR, PB terminal repeats; B1/B2, post-Gateway cloning sites. (b) Stable dox-independent cell lines activate alkaline phosphatase (AP), SSEA1 and Nanog. Representative images are from a single cell line (1B). (c) Dox regulation of PB-delivered factors as monitored by transgene-specific RT-PCR analysis (vector-based reverse primer). Reprogrammed cell lines and controls were grown in the presence (+) or absence (−) of dox for 2 passages (~96 hrs). The induction (Oct4, Sox2) or maintenance (c-Myc, Klf4) of endogenous gene expression was determined by RT-PCR using 3′UTR-directed reverse primers. (d) Residual transgene expression in reprogrammed cell lines visualized as lacZ activity from the transcription-linked βgeo reporter gene (Fig. 1a). The level and mosaicism of lacZ after dox withdrawal correlates roughly with basal level transcription detected by RT-PCR. Scale bars are 200 μm.
Figure 2
Seamless factor removal from iPS cells using transposase-stimulated PB excision. (a) Schematic of the MKOS-containing PB-TET transposon. (b) Genomic integration site of the individual transposons in scB1 and scC5 lines. Capital letters represent flanking genomic sequences while lower case letters are transposon TR sequences. (c) Sequence analyses revealed that no mutation was left behind following transposon-mediated removal in the majority of sublines (10 of 11). One single C5 subclone harbored a TTAA duplication at the excision site. (d) Molecular demonstration of transposon removal in representative subclones. From top to bottom: GFP PCR ensures all cell lines are derivatives of rtTA MEFs (R1 ES negative control); βgeo PCR detects the presence of transposons regardless of genomic location; Chr11- and Chr16-specific PCR across the TTAA tetranucleotide insertion site. scB1 and scC5 are hemizygous for the transposon and amplify the wild type allele; 5′ and 3′ junction PCR; 3-primer PCR for the wild type allele and the transposon-genome junction. (e) RT-PCR analysis of the single transposon induced iPS cell lines and their factor-removed derivatives reveals maintenance of hallmark pluripotency gene expression. R1 ES cells and parental rtTA-MEFs serve as positive and negative controls, respectively.
Figure 3
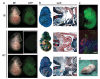
Cell lines reprogrammed by PB-mediated factor transposition are pluripotent. (a) Contribution of PB-TET cell lines to embryonic development. Chimaeras dissected at 10.5dpc with the strongest contribution of PB-iPS-derived cells were easily detected as GFP positive (BF - bright field). (b) Whole mount lacZ staining of chimaeric embryos and their sections show contribution of PB-iPS lacZ positive cells to derivatives of all three embryonic germ layers. Red arrowhead – neural tube (ectoderm); yellow arrowhead – dorsal aorta (mesoderm); black arrow foregut epithelium (endoderm). (c) Completely iPS cell-derived (1B) 13.5 dpc embryo generated by tetraploid embryo complementation. Immunohistochemistry on sections through its genital ridge shows the iPS cell contribution to germ cells (Vasa+, red). (d) Adult chimaera obtained by aggregating 1B PB-iPS cells with diploid eight cell stage ICR (albino) embryos. Scale bars are 100 μm.
Figure 4
Properties of secondary fibroblast (2°F) reprogramming. FACS analysis establishing the dynamics of SSEA1 activation in 2°F/1B and 2°F/6C cell lines. The inset shows colony formation as early as d5. On d6 of reprogramming gene induction, the cultures were also passed by standard trypsinization and analyzed in parallel to eliminate the negative effect of cell overgrowth.
Comment in
- Stem cells: Low-risk reprogramming.
Pera MF. Pera MF. Nature. 2009 Apr 9;458(7239):715-6. doi: 10.1038/458715a. Nature. 2009. PMID: 19360075 No abstract available.
Similar articles
- Virus-free induction of pluripotency and subsequent excision of reprogramming factors.
Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Kaji K, et al. Nature. 2009 Apr 9;458(7239):771-5. doi: 10.1038/nature07864. Epub 2009 Mar 1. Nature. 2009. PMID: 19252477 Free PMC article. - Non-viral reprogramming of fibroblasts into induced pluripotent stem cells by Sleeping Beauty and piggyBac transposons.
Talluri TR, Kumar D, Glage S, Garrels W, Ivics Z, Debowski K, Behr R, Kues WA. Talluri TR, et al. Biochem Biophys Res Commun. 2014 Jul 18;450(1):581-7. doi: 10.1016/j.bbrc.2014.06.014. Epub 2014 Jun 10. Biochem Biophys Res Commun. 2014. PMID: 24928388 - Reprogramming of murine and human somatic cells using a single polycistronic vector.
Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Carey BW, et al. Proc Natl Acad Sci U S A. 2009 Jan 6;106(1):157-62. doi: 10.1073/pnas.0811426106. Epub 2008 Dec 24. Proc Natl Acad Sci U S A. 2009. PMID: 19109433 Free PMC article. - Advances in reprogramming somatic cells to induced pluripotent stem cells.
Patel M, Yang S. Patel M, et al. Stem Cell Rev Rep. 2010 Sep;6(3):367-80. doi: 10.1007/s12015-010-9123-8. Stem Cell Rev Rep. 2010. PMID: 20336395 Free PMC article. Review. - PiggyBac toolbox.
Di Matteo M, Mátrai J, Belay E, Firdissa T, Vandendriessche T, Chuah MK. Di Matteo M, et al. Methods Mol Biol. 2012;859:241-54. doi: 10.1007/978-1-61779-603-6_14. Methods Mol Biol. 2012. PMID: 22367876 Review.
Cited by
- How microRNAs facilitate reprogramming to pluripotency.
Anokye-Danso F, Snitow M, Morrisey EE. Anokye-Danso F, et al. J Cell Sci. 2012 Sep 15;125(Pt 18):4179-87. doi: 10.1242/jcs.095968. Epub 2012 Oct 17. J Cell Sci. 2012. PMID: 23077173 Free PMC article. Review. - Generation of virus-free induced pluripotent stem cell clones on a synthetic matrix via a single cell subcloning in the naïve state.
Nishishita N, Shikamura M, Takenaka C, Takada N, Fusaki N, Kawamata S. Nishishita N, et al. PLoS One. 2012;7(6):e38389. doi: 10.1371/journal.pone.0038389. Epub 2012 Jun 13. PLoS One. 2012. PMID: 22719883 Free PMC article. - Generation of mouse and human induced pluripotent stem cells (iPSC) from primary somatic cells.
Lorenzo IM, Fleischer A, Bachiller D. Lorenzo IM, et al. Stem Cell Rev Rep. 2013 Aug;9(4):435-50. doi: 10.1007/s12015-012-9412-5. Stem Cell Rev Rep. 2013. PMID: 23104133 - Induced pluripotent stem cells and personalized medicine: current progress and future perspectives.
Chun YS, Byun K, Lee B. Chun YS, et al. Anat Cell Biol. 2011 Dec;44(4):245-55. doi: 10.5115/acb.2011.44.4.245. Epub 2011 Dec 30. Anat Cell Biol. 2011. PMID: 22254153 Free PMC article. - Effect of Honokiol on culture time and survival of Alzheimer's disease iPSC-derived neurons.
Le DTT, Vu CM, Ly TTB, Nguyen NT, Nguyen PTM, Chu HH. Le DTT, et al. Bioimpacts. 2024;14(1):27652. doi: 10.34172/bi.2023.27652. Epub 2023 Aug 5. Bioimpacts. 2024. PMID: 38327632 Free PMC article.
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. - PubMed
- Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1(1):55–70. - PubMed
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. - PubMed
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25(10):1177–1181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials