Characterization of mouse IFT complex B - PubMed (original) (raw)
Characterization of mouse IFT complex B
John A Follit et al. Cell Motil Cytoskeleton. 2009 Aug.
Abstract
The primary cilium plays a key role in the development of mammals and in the maintenance of health. Primary cilia are assembled and maintained by the process of intraflagellar transport (IFT). In this work, we characterize mouse IFT complex B by identifying all of the mammalian orthologues of complex B and B-associated proteins previously identified in Chlamydomonas and Caenorhabditis and also identify a new component (IFT25/Hspb11) of complex B by database analysis. We tagged each of these proteins with the FLAG epitope and show that all except IFT172 and IFT20 localize to cilia and the peri-basal body or centrosomal region at the base of cilia. All of the proteins except IFT172 immunoprecipitate IFT88 indicating that they are co-assembled into a complex. IFT20 is the only complex B protein that localizes to the Golgi apparatus. However, overexpression of IFT54/Traf3ip1, the mouse orthologue of Dyf-11/Elipsa, displaces IFT20 from the Golgi apparatus. IFT54 does not localize to the Golgi complex nor does it interact with GMAP210, which is the protein that anchors IFT20 to the Golgi apparatus. This suggests that IFT54s effect on IFT20 is a dominant negative phenotype caused by its overexpression. Cell Motil. Cytoskeleton 2009. (c) 2009 Wiley-Liss, Inc.
Figures
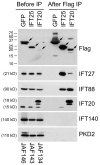
Figure 1. C1orf41/IFT25 is a subunit of IFT complex B
Mouse IMCD3 cells stably transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed vertically at the top of the figure while name of the expression construct is listed vertically at the bottom. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Abs used for the western blots are listed on the right side. The western blot of IFT20 shows endogenous IFT20 at 15 kD and FLAG-IFT20 at 18 kD along with a degradation product of the FLAG-IFT20 that runs slightly faster. Note that both IFT20 and C1orf41/IFT25 precipitated complex B subunits (IFT27, IFT88, IFT20) but not complex A (IFT140) or control proteins (PKD2).
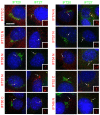
Figure 2. Localization of complex B proteins in mouse kidney cells
Mouse IMCD3 cells stably transfected with FLAG expression constructs were serum starved for 48 hr, fixed with paraformaldehyde and stained with DAPI (blue), FLAG (red) and IFT20 (green, left panel in each pair) or IFT27 (green, right panel in each pair). The transfected FLAG construct is listed on the left side of each set of panels. N and C refer the position of the tags. Cilia are marked with arrows. Insets are the FLAG (red) channels of the cilia to more clearly document the localization of the FLAG-tagged protein to this organelle. Note that all proteins except IFT172 (with either an N- or C-terminal tag), IFT20 and GFP localize to cilia. Also note that IFT20 is the only protein localized to the Golgi complex and that IFT54 displaces native IFT20 from the Golgi complex. Size bar is 10 μm and relevant for all panels.
Figure 2. Localization of complex B proteins in mouse kidney cells
Mouse IMCD3 cells stably transfected with FLAG expression constructs were serum starved for 48 hr, fixed with paraformaldehyde and stained with DAPI (blue), FLAG (red) and IFT20 (green, left panel in each pair) or IFT27 (green, right panel in each pair). The transfected FLAG construct is listed on the left side of each set of panels. N and C refer the position of the tags. Cilia are marked with arrows. Insets are the FLAG (red) channels of the cilia to more clearly document the localization of the FLAG-tagged protein to this organelle. Note that all proteins except IFT172 (with either an N- or C-terminal tag), IFT20 and GFP localize to cilia. Also note that IFT20 is the only protein localized to the Golgi complex and that IFT54 displaces native IFT20 from the Golgi complex. Size bar is 10 μm and relevant for all panels.
Figure 3. Immunoprecipitation of FLAG-tagged proteins
Mouse IMCD3 cells stably transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed at the top of the figure. Abs used for the western blots are listed on the right side. Arrows indicate the FLAG-tagged protein. * at ~37 kD marks a non-specific protein that is immunoprecipitated by the FLAG antibody. Ladders are shown to the left of each of the gels. The sizes of the markers are the same for all and shown on the left most ladder. Input (left most lane of the first gel) is starting material from the IFT20 immunoprecipitation equivalent to 1/6 of the amount in the precipitated lane. Note that GFP-FLAG was much more abundant than the other proteins and 1/5 as much of this immunoprecipitation was loaded on the gel for the FLAG western. The amount was not altered in the gels for the IFT88, GMAP210 or IFT140 westerns.
Figure 4. Interaction between IFT54 and the IFT complex
Mouse IMCD3 cells transiently transfected with FLAG expression constructs were lysed 48 hrs after electroporation, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed vertically at the top of the figure while name of the expression construct is listed vertically at the bottom. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Note that GMAP210 is less abundant than the other proteins and is not visible at exposures reasonable for them. The right most lane (*) shows a longer exposure of immunoprecipitated GMAP210 from a different gel. Abs used for the western blots are listed on the right side. The western blot of IFT20 shows endogenous IFT20 at 15 kD and FLAG-IFT20 at 18 kD along with a degradation product of the FLAG-IFT20 that runs slightly faster. Note that the blank lane was inadvertently left out of the Rabaptin5 gel. To allow the lanes to line up, the image was split in the middle.
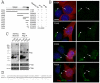
Figure 5. Characterization of the IFT20 binding site on IFT54
A. Diagram summarizing data from experiments to map the IFT20 binding site on IFT54. Map illustrating the motif structure of IFT54 is shown on the top. Expression construct names are on the left. Exact amino acids are listed in the materials and methods. Arg rich domain is an unusual stretch of alternating positive and negatively charged residues. The coiled-coil domain is marked by series of circles. The IFT20 binding domain is marked by a solid dark bar. Summary of the activity of each of the constructs is shown on the right. B. IF of selected constructs to illustrate the ability of the C-terminal end of IFT54 (Red) to displace native IFT20 (green) from the Golgi complex. Left panel is a composite of red and green channels while the right panel shows only IFT20 staining in green. Arrows mark transfected cells. Arrowheads mark untransfected cells. Scale bar is 10 μm and relevant for all panels. C. Immunoprecipitation of selected constructs to illustrate the ability of C-terminal fragments of IFT54 to precipitate IFT20 but not IFT88. Mouse IMCD3 cells transiently transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged expression construct is listed at the top of the figure. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Antibodies used for western blotting are listed on the right. D. ClustalW alignment of the IFT20 binding site from IFT54 to a portion of the IFT20 binding site from GMAP210. * indicate identity, : and . mark similarity.
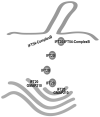
Figure 6. Model for the coordination of IFT20 in the Golgi IFT complex and complex B
The finding that IFT20 is part of the Golgi IFT complex as well as complex B presents interesting possibilities for its function. This speculative model presents our view of how this may be occurring. IFT20 is anchored to the Golgi membrane by the golgin protein GMAP210 [Follit et al., 2008]. IFT20 is dynamic and moves from the Golgi complex to the cilium [Follit et al., 2006] but GMAP210 appears to remain at the Golgi. IFT20 binds directly to the complex B subunit IFT54 [this work and Omori et al. 2008] but IFT54 does not appear to localize to the Golgi complex and it is not part of the Golgi IFT complex [this work]. Thus, we hypothesize that IFT20 and GMAP210 function at the Golgi complex to sort proteins into vesicles destined for the ciliary membrane. These vesicles leave the Golgi with IFT20 associated with them. Once the vesicles arrive at the base of the cilium, IFT20 on the vesicles interacts with the IFT54 subunit of complex B to form the complete IFT complex. The vesicles fuse with the plasma membrane at the base of the cilium and the IFT complex with attached cargos is carried into the cilium by kinesin-2. By having the IFT complex assembled on the surface of the vesicle, the transport of membrane and non-membranous cargos could be coordinated.
Similar articles
- The Golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex.
Follit JA, San Agustin JT, Xu F, Jonassen JA, Samtani R, Lo CW, Pazour GJ. Follit JA, et al. PLoS Genet. 2008 Dec;4(12):e1000315. doi: 10.1371/journal.pgen.1000315. Epub 2008 Dec 26. PLoS Genet. 2008. PMID: 19112494 Free PMC article. - The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly.
Follit JA, Tuft RA, Fogarty KE, Pazour GJ. Follit JA, et al. Mol Biol Cell. 2006 Sep;17(9):3781-92. doi: 10.1091/mbc.e06-02-0133. Epub 2006 Jun 14. Mol Biol Cell. 2006. PMID: 16775004 Free PMC article. - IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia.
Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. Baker SA, et al. J Biol Chem. 2003 Sep 5;278(36):34211-8. doi: 10.1074/jbc.M300156200. Epub 2003 Jun 23. J Biol Chem. 2003. PMID: 12821668 - Intraflagellar transport 20 cilia-dependent and cilia-independent signaling pathways in cell development and tissue homeostasis.
Jin FC, Zhou MH, Chen JJ, Lin Y, Zhang QW, Xu QX, Zhang CC, Zhang ZG. Jin FC, et al. Int J Dev Biol. 2022;66(4-5-6):333-347. doi: 10.1387/ijdb.220072fj. Int J Dev Biol. 2022. PMID: 35980193 Review. - Intraflagellar transport 20: New target for the treatment of ciliopathies.
Zhou MH, Lin Y, Zhang ZG. Zhou MH, et al. Biochim Biophys Acta Mol Cell Res. 2020 Jul;1867(7):118641. doi: 10.1016/j.bbamcr.2019.118641. Epub 2019 Dec 30. Biochim Biophys Acta Mol Cell Res. 2020. PMID: 31893523 Review.
Cited by
- Intraflagellar transport complex structure and cargo interactions.
Bhogaraju S, Engel BD, Lorentzen E. Bhogaraju S, et al. Cilia. 2013 Aug 14;2(1):10. doi: 10.1186/2046-2530-2-10. Cilia. 2013. PMID: 23945166 Free PMC article. - Distinct functions for IFT140 and IFT20 in opsin transport.
Crouse JA, Lopes VS, Sanagustin JT, Keady BT, Williams DS, Pazour GJ. Crouse JA, et al. Cytoskeleton (Hoboken). 2014 May;71(5):302-10. doi: 10.1002/cm.21173. Epub 2014 Mar 25. Cytoskeleton (Hoboken). 2014. PMID: 24619649 Free PMC article. - Chlamydomonas LZTFL1 mediates phototaxis via controlling BBSome recruitment to the basal body and its reassembly at the ciliary tip.
Sun WY, Xue B, Liu YX, Zhang RK, Li RC, Xin W, Wu M, Fan ZC. Sun WY, et al. Proc Natl Acad Sci U S A. 2021 Aug 31;118(35):e2101590118. doi: 10.1073/pnas.2101590118. Proc Natl Acad Sci U S A. 2021. PMID: 34446551 Free PMC article. - Identification of the TTC26 Splice Variant in a Novel Complex Ciliopathy Syndrome with Biliary, Renal, Neurological, and Skeletal Manifestations.
Alfadhel M, Umair M, Almuzzaini B, Asiri A, Al Tuwaijri A, Alhamoudi K, Alyafee Y, Al-Owain M. Alfadhel M, et al. Mol Syndromol. 2021 Jun;12(3):133-140. doi: 10.1159/000513829. Epub 2021 May 11. Mol Syndromol. 2021. PMID: 34177428 Free PMC article. - Overall Architecture of the Intraflagellar Transport (IFT)-B Complex Containing Cluap1/IFT38 as an Essential Component of the IFT-B Peripheral Subcomplex.
Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. Katoh Y, et al. J Biol Chem. 2016 May 20;291(21):10962-75. doi: 10.1074/jbc.M116.713883. Epub 2016 Mar 15. J Biol Chem. 2016. PMID: 26980730 Free PMC article.
References
- Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem. 2003;278:34211–34218. - PubMed
- Bellyei S, Szigeti A, Boronkai A, Pozsgai E, Gomori E, Melegh B, Janaky T, Bognar Z, Hocsak E, Sumegi B, Gallyas F., Jr. Inhibition of cell death by a novel 16.2 kD heat shock protein predominantly via Hsp90 mediated lipid rafts stabilization and Akt activation pathway. Apoptosis. 2007a;12:97–112. - PubMed
- Bellyei S, Szigeti A, Pozsgai E, Boronkai A, Gomori E, Hocsak E, Farkas R, Sumegi B, Gallyas F., Jr. Preventing apoptotic cell death by a novel small heat shock protein. Eur J Cell Biol. 2007b;86:161–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK032520/DK/NIDDK NIH HHS/United States
- R01 GM060992-09/GM/NIGMS NIH HHS/United States
- R01 GM060992/GM/NIGMS NIH HHS/United States
- GM060992/GM/NIGMS NIH HHS/United States
- R01 GM060992-08/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases