The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis - PubMed (original) (raw)
. 2009 Mar 17;106(11):4549-54.
doi: 10.1073/pnas.0810280106. Epub 2009 Mar 2.
Peter D Jones, Laurel Russell, Anne Medhurst, Susana Ubeda Tomás, Prabhavathi Talloji, Julietta Marquez, Heike Schmuths, Swee-Ang Tung, Ian Taylor, Steven Footitt, Andreas Bachmair, Frederica L Theodoulou, Michael J Holdsworth
Affiliations
- PMID: 19255443
- PMCID: PMC2649959
- DOI: 10.1073/pnas.0810280106
The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis
Tara J Holman et al. Proc Natl Acad Sci U S A. 2009.
Abstract
The N-end rule pathway targets protein degradation through the identity of the amino-terminal residue of specific protein substrates. Two components of this pathway in Arabidopsis thaliana, PROTEOLYSIS6 (PRT6) and arginyl-tRNA:protein arginyltransferase (ATE), were shown to regulate seed after-ripening, seedling sugar sensitivity, seedling lipid breakdown, and abscisic acid (ABA) sensitivity of germination. Sensitivity of prt6 mutant seeds to ABA inhibition of endosperm rupture reduced with after-ripening time, suggesting that seeds display a previously undescribed window of sensitivity to ABA. Reduced root growth of prt6 alleles and the ate1 ate2 double mutant was rescued by exogenous sucrose, and the breakdown of lipid bodies and seed-derived triacylglycerol was impaired in mutant seedlings, implicating the N-end rule pathway in control of seed oil mobilization. Epistasis analysis indicated that PRT6 control of germination and establishment, as exemplified by ABA and sugar sensitivity, as well as storage oil mobilization, occurs at least in part via transcription factors ABI3 and ABI5. The N-end rule pathway of protein turnover is therefore postulated to inactivate as-yet unidentified key component(s) of ABA signaling to influence the seed-to-seedling transition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
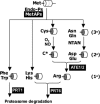
Fig. 1.
Diagrammatic representation of the N-end rule pathway associated with PRT6 function. Components for which orthologous Arabidopsis genes have been identified are highlighted. Protein substrate (represented as a cylinder) is cleaved by a methionine aminopeptidase (MetAP) or a specific endopeptidase (Endo-P) at a unique amino acid sequence (dotted curve). Cleavage reveals destabilizing amino acid residues, which may be further modified by enzymatic (NTAN, amino-terminal amidohydrolase; ATE1/2, Arg-tRNA-protein transferase) or chemical (O2, NO) action. 1°, 2°, and 3° indicate primary, secondary, and tertiary destabilizing amino-terminal residues, respectively. Generation of a primary destabilizing residue (1°) leads to targeting by E3 ubiquitin ligases PROTEOLYSIS6 (PRT6) or PRT1, depending on the nature of the destabilizing residue. C*, oxidized derivative of cysteine.
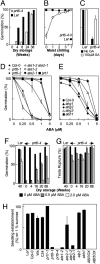
Fig. 2.
ABA sensitivity of germination, after-ripening, and sucrose sensitivity of establishment. (A) After-ripening of WT (L_er_) and prt6-4 seeds assayed on water agarose after 7 days. (B) Germination potential of seeds imbibed for 7 days on water-agarose media after increasing periods of cold, moist chilling. (C) Germination potential of seeds imbibed for 7 days on agarose media containing 1/2MS in the absence (black bars) and presence (white bars) of 100 μM GA3. (D and E) Germination potential after 7 days of WT, mutant, and transgenic seeds assayed on 1/2MS media in the presence of exogenous ABA following 2 days of moist chilling. Seeds were stored for 2 months before assay. Respective WT and mutant seeds are: Col-0, prt1-1, prt6-1, prt6-2, ate1-2, ate2-1, abh1, ahg1, 2, 3 (3–5 and 36–39). Data points for ate2-1 are slightly obscured by those of prt1. Results obtained for ABI3OX, ABI5OX, and aip2 are presented in
Fig. S2_D_
. (F and G) Change in sensitivity of endosperm rupture (F) and testa rupture (G) to exogenous ABA, with time of dry storage of cold, chilled WT (L_er_) and prt6-4 seeds, assayed on 1/2MS media and measured at 7 days following 2 days of moist chilling. (H) Sucrose sensitivity of establishment (greening of cotyledons) of WT, transgenic, and mutant seedlings grown on water-agarose supplemented with 1% (wt/vol) sucrose measured at 7 days, following 2 days of moist chilling. Dose–response curves are presented in
Fig. S4
. Data represent means ± SE of the mean.
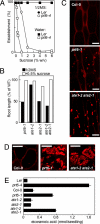
Fig. 3.
Influence of PRT6 and ATE1/2 on lipid degradation during establishment. (A) Establishment of prt6-4 and WT (L_er_) seedlings after 7 days of growth on increasing concentrations of sucrose either in the presence (1/2MS) or absence (water) of 1/2 MS following 2 days of moist chilling. (B) Influence of exogenous sucrose on root length of prt6 and ate1 and ate2 alleles in relation to WT, measured at 7 days of imbibition on 1/2MS media following 2 days of moist chilling. (C and D) Representative confocal microscopy images of hypocotyl epidermal (C) and endosperm (D) cells (5 days of imbibition on 1/2MS, no added sucrose, following 2 days of moist chilling) stained with Nile Red to reveal oil bodies, which can be seen as red-staining spherical inclusions in prt6-1 and ate1 ate2, but not in Col-0. (Scale bar: 20 μm.) Brightfield images are presented in
Fig. S5
. (E) Eicosenoic acid (20:1) content of 5-day-old seedlings grown on 1/2MS media following 2 days of moist chilling. Data represent means ± SE of the mean.
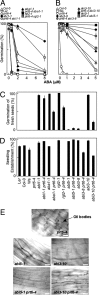
Fig. 4.
Genetic interactions of PRT6 with components of ABA and GA biosynthesis and signaling. (A and B) Germination potential after 7 days of WT mutant and transgenic seeds on 1/2MS media in the presence of exogenous ABA, following 2 days of moist chilling. abi1-1 (L_er_ background) (26), aba1-1 (L_er_) (25), rgl2-1 (L_er_) (24), abi3-8 (Col-0), abi3-10 (Col-0) (28), abi5-1 (Ws) (27). (C) Germination potential, after 7 days, of freshly harvested seeds on water-agarose. (D) Seedling establishment, after 7 days, on water-agarose containing 1% (wt/vol) sucrose, following 2 days of moist chilling (dose–response curves are presented in
Fig. S4
.). (E) Light micrographs of hypocotyl epidermis of single- and double-mutant seeds showing retention of lipid bodies in prt6-4, but not in single- or double-mutant combinations with abi3-10 or abi5-1, on 1/2MS media after 5 days, following 2 days of moist chilling. (Scale bar: 20 μm.). Data represent means ± SE of the mean.
Similar articles
- CONSTITUTIVELY PHOTOMORPHOGENIC1 promotes ABA-mediated inhibition of post-germination seedling establishment.
Yadukrishnan P, Rahul PV, Ravindran N, Bursch K, Johansson H, Datta S. Yadukrishnan P, et al. Plant J. 2020 Jul;103(2):481-496. doi: 10.1111/tpj.14844. Epub 2020 Jun 17. Plant J. 2020. PMID: 32436306 - Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination.
Pan J, Wang H, Hu Y, Yu D. Pan J, et al. Plant J. 2018 Aug;95(3):529-544. doi: 10.1111/tpj.13969. Epub 2018 Jun 8. Plant J. 2018. PMID: 29771466 - The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development.
Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D, Li C. Bu Q, et al. Plant Physiol. 2009 May;150(1):463-81. doi: 10.1104/pp.109.135269. Epub 2009 Mar 13. Plant Physiol. 2009. PMID: 19286935 Free PMC article. - Genetic interactions between ABA signalling and the Arg/N-end rule pathway during Arabidopsis seedling establishment.
Zhang H, Gannon L, Jones PD, Rundle CA, Hassall KL, Gibbs DJ, Holdsworth MJ, Theodoulou FL. Zhang H, et al. Sci Rep. 2018 Oct 12;8(1):15192. doi: 10.1038/s41598-018-33630-5. Sci Rep. 2018. PMID: 30315202 Free PMC article. - Light and abscisic acid interplay in early seedling development.
Yadukrishnan P, Datta S. Yadukrishnan P, et al. New Phytol. 2021 Jan;229(2):763-769. doi: 10.1111/nph.16963. Epub 2020 Oct 25. New Phytol. 2021. PMID: 32984965 Review.
Cited by
- The Structure of Saccharomyces cerevisiae Arginyltransferase 1 (ATE1).
Van V, Ejimogu NE, Bui TS, Smith AT. Van V, et al. J Mol Biol. 2022 Nov 15;434(21):167816. doi: 10.1016/j.jmb.2022.167816. Epub 2022 Sep 8. J Mol Biol. 2022. PMID: 36087779 Free PMC article. - Identification of Targets and Interaction Partners of Arginyl-tRNA Protein Transferase in the Moss Physcomitrella patens.
Hoernstein SN, Mueller SJ, Fiedler K, Schuelke M, Vanselow JT, Schuessele C, Lang D, Nitschke R, Igloi GL, Schlosser A, Reski R. Hoernstein SN, et al. Mol Cell Proteomics. 2016 Jun;15(6):1808-22. doi: 10.1074/mcp.M115.057190. Epub 2016 Apr 11. Mol Cell Proteomics. 2016. PMID: 27067052 Free PMC article. - Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis.
Sasidharan R, Mustroph A. Sasidharan R, et al. Plant Cell. 2011 Dec;23(12):4173-83. doi: 10.1105/tpc.111.093880. Epub 2011 Dec 29. Plant Cell. 2011. PMID: 22207573 Free PMC article. - An analysis of dormancy, ABA responsiveness, after-ripening and pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) caryopses.
Gerjets T, Scholefield D, Foulkes MJ, Lenton JR, Holdsworth MJ. Gerjets T, et al. J Exp Bot. 2010;61(2):597-607. doi: 10.1093/jxb/erp329. Epub 2009 Nov 18. J Exp Bot. 2010. PMID: 19923197 Free PMC article. - Biochemical analysis of protein arginylation.
Wang J, Yates JR 3rd, Kashina A. Wang J, et al. Methods Enzymol. 2019;626:89-113. doi: 10.1016/bs.mie.2019.07.028. Epub 2019 Aug 1. Methods Enzymol. 2019. PMID: 31606094 Free PMC article.
References
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. - PubMed
- Tasaki T, Kwon YT. The mammalian N-end rule pathway: New insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. - PubMed
- Garzon M, et al. PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 2007;581:3189–3196. - PubMed
- Yoshida S, Ito M, Callis J, Nishida I, Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA: Protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials