Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance - PubMed (original) (raw)
Review
Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance
Lauren S Waters et al. Microbiol Mol Biol Rev. 2009 Mar.
Abstract
DNA repair and DNA damage tolerance machineries are crucial to overcome the vast array of DNA damage that a cell encounters during its lifetime. In this review, we summarize the current state of knowledge about the eukaryotic DNA damage tolerance pathway translesion synthesis (TLS), a process in which specialized DNA polymerases replicate across from DNA lesions. TLS aids in resistance to DNA damage, presumably by restarting stalled replication forks or filling in gaps that remain in the genome due to the presence of DNA lesions. One consequence of this process is the potential risk of introducing mutations. Given the role of these translesion polymerases in mutagenesis, we discuss the significant regulatory mechanisms that control the five known eukaryotic translesion polymerases: Rev1, Pol zeta, Pol kappa, Pol eta, and Pol iota.
Figures
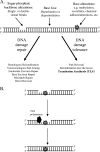
FIG. 1.
DNA damage repair and bypass mechanisms. (A) DNA damage results in breakage of the sugar-phosphate backbone, DNA base loss (indicated by a gap in the DNA), or base alterations (as indicated by the gray star). This damage can be repaired/removed from the DNA strand or tolerated, in which case the DNA lesion remains but cellular processes continue. (B) An example of the DNA damage tolerance mechanism TLS, whereby a damaged DNA template is replicated using a TLS polymerase and the damage remains in the genome. A more detailed mechanism of TLS is described in the text and represented in Fig. 4.
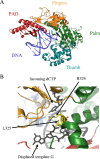
FIG. 2.
Crystal structures of two Y family polymerases. (A) Cocrystal structure of the S. cerevisiae TLS Pol η with a DNA template containing a cisplatin cross-link. The structure is oriented to highlight the right-hand architecture as seen in both TLS and replicative polymerases. (Based on data from reference and the RSCB protein data bank [PDB ID number 2r8j].) (B) Close-up view of the unique lesion bypass mechanism of Rev1 from S. cerevisiae. Highlighted are the novel leucine (L325) that helps to flip out the template guanine and the catalytic arginine (R324) that hydrogen bonds to stabilize the incoming dCTP. The domains of Rev1 are colored as in panel A, with the exception of the DNA, which is shown in black. (Based on data from reference and the RCSB protein data bank [PDB ID number 2aq4].)
FIG. 3.
Cartoon representation of the protein domains in the human B family TLS polymerase ζ and the Y family TLS polymerases Rev1, κ, ι, and η. aa, amino acids. (Based on data from references and .)
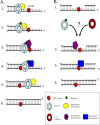
FIG. 4.
Two nonexclusive models for TLS: the polymerase-switching model (A) and the gap-filling model (B). See text for details.
Similar articles
- Filling gaps in translesion DNA synthesis in human cells.
Quinet A, Lerner LK, Martins DJ, Menck CFM. Quinet A, et al. Mutat Res Genet Toxicol Environ Mutagen. 2018 Dec;836(Pt B):127-142. doi: 10.1016/j.mrgentox.2018.02.004. Epub 2018 Feb 23. Mutat Res Genet Toxicol Environ Mutagen. 2018. PMID: 30442338 Review. - Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric polymerase (Pol) ζ, and Pol κ.
Wojtaszek J, Lee CJ, D'Souza S, Minesinger B, Kim H, D'Andrea AD, Walker GC, Zhou P. Wojtaszek J, et al. J Biol Chem. 2012 Sep 28;287(40):33836-46. doi: 10.1074/jbc.M112.394841. Epub 2012 Aug 2. J Biol Chem. 2012. PMID: 22859295 Free PMC article. - Translesion Synthesis of the N(2)-2'-Deoxyguanosine Adduct of the Dietary Mutagen IQ in Human Cells: Error-Free Replication by DNA Polymerase κ and Mutagenic Bypass by DNA Polymerases η, ζ, and Rev1.
Bose A, Millsap AD, DeLeon A, Rizzo CJ, Basu AK. Bose A, et al. Chem Res Toxicol. 2016 Sep 19;29(9):1549-59. doi: 10.1021/acs.chemrestox.6b00221. Epub 2016 Aug 17. Chem Res Toxicol. 2016. PMID: 27490094 Free PMC article. - Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis.
Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Guo C, et al. EMBO J. 2003 Dec 15;22(24):6621-30. doi: 10.1093/emboj/cdg626. EMBO J. 2003. PMID: 14657033 Free PMC article. - Eukaryotic TLS polymerases.
Tomczyk P, Synowiec E, Wysokiński D, Woźniak K. Tomczyk P, et al. Postepy Hig Med Dosw (Online). 2016 May 21;70(0):522-33. doi: 10.5604/17322693.1202481. Postepy Hig Med Dosw (Online). 2016. PMID: 27333922 Review.
Cited by
- Leveraging the replication stress response to optimize cancer therapy.
Cybulla E, Vindigni A. Cybulla E, et al. Nat Rev Cancer. 2023 Jan;23(1):6-24. doi: 10.1038/s41568-022-00518-6. Epub 2022 Nov 2. Nat Rev Cancer. 2023. PMID: 36323800 Free PMC article. Review. - HUWE1 interacts with PCNA to alleviate replication stress.
Choe KN, Nicolae CM, Constantin D, Imamura Kawasawa Y, Delgado-Diaz MR, De S, Freire R, Smits VA, Moldovan GL. Choe KN, et al. EMBO Rep. 2016 Jun;17(6):874-86. doi: 10.15252/embr.201541685. Epub 2016 May 4. EMBO Rep. 2016. PMID: 27146073 Free PMC article. - Human DNA Polymerase ν Catalyzes Correct and Incorrect DNA Synthesis with High Catalytic Efficiency.
Gowda AS, Moldovan GL, Spratt TE. Gowda AS, et al. J Biol Chem. 2015 Jun 26;290(26):16292-303. doi: 10.1074/jbc.M115.653287. Epub 2015 May 11. J Biol Chem. 2015. PMID: 25963146 Free PMC article. - Analysis of CPD ultraviolet lesion bypass in chicken DT40 cells: polymerase η and PCNA ubiquitylation play identical roles.
Varga A, Marcus AP, Himoto M, Iwai S, Szüts D. Varga A, et al. PLoS One. 2012;7(12):e52472. doi: 10.1371/journal.pone.0052472. Epub 2012 Dec 18. PLoS One. 2012. PMID: 23272247 Free PMC article. - Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining.
Kent T, Mateos-Gomez PA, Sfeir A, Pomerantz RT. Kent T, et al. Elife. 2016 Jun 17;5:e13740. doi: 10.7554/eLife.13740. Elife. 2016. PMID: 27311885 Free PMC article.
References
- Albertella, M. R., C. M. Green, A. R. Lehmann, and M. J. O'Connor. 2005. A role for polymerase eta in the cellular tolerance to cisplatin-induced damage. Cancer Res. 659799-9806. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5-F32-GM078966/GM/NIGMS NIH HHS/United States
- R01 ES015818/ES/NIEHS NIH HHS/United States
- P30 ES002109/ES/NIEHS NIH HHS/United States
- 5-R01-ES015818/ES/NIEHS NIH HHS/United States
- F32 GM078966/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases