Ultrafast and memory-efficient alignment of short DNA sequences to the human genome - PubMed (original) (raw)
Ultrafast and memory-efficient alignment of short DNA sequences to the human genome
Ben Langmead et al. Genome Biol. 2009.
Abstract
Bowtie is an ultrafast, memory-efficient alignment program for aligning short DNA sequence reads to large genomes. For the human genome, Burrows-Wheeler indexing allows Bowtie to align more than 25 million reads per CPU hour with a memory footprint of approximately 1.3 gigabytes. Bowtie extends previous Burrows-Wheeler techniques with a novel quality-aware backtracking algorithm that permits mismatches. Multiple processor cores can be used simultaneously to achieve even greater alignment speeds. Bowtie is open source (http://bowtie.cbcb.umd.edu).
Figures
Figure 1
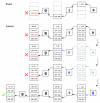
Burrows-Wheeler transform. (a) The Burrows-Wheeler matrix and transformation for 'acaacg'. (b) Steps taken by EXACTMATCH to identify the range of rows, and thus the set of reference suffixes, prefixed by 'aac'. (c) UNPERMUTE repeatedly applies the last first (LF) mapping to recover the original text (in red on the top line) from the Burrows-Wheeler transform (in black in the rightmost column).
Figure 2
Exact matching versus inexact alignment. Illustration of how EXACTMATCH (top) and Bowtie's aligner (bottom) proceed when there is no exact match for query 'ggta' but there is a one-mismatch alignment when 'a' is replaced by 'g'. Boxed pairs of numbers denote ranges of matrix rows beginning with the suffix observed up to that point. A red X marks where the algorithm encounters an empty range and either aborts (as in EXACTMATCH) or backtracks (as in the inexact algorithm). A green check marks where the algorithm finds a nonempty range delimiting one or more occurrences of a reportable alignment for the query.
Figure 3
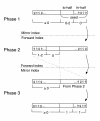
The three phases of the Bowtie algorithm for the Maq-like policy. A three-phase approach finds alignments for two-mismatch cases 1 to 4 while minimizing backtracking. Phase 1 uses the mirror index and invokes the aligner to find alignments for cases 1 and 2. Phases 2 and 3 cooperate to find alignments for case 3: Phase 2 finds partial alignments with mismatches only in the hi-half, and phase 3 attempts to extend those partial alignments into full alignments. Finally, phase 3 invokes the aligner to find alignments for case 4.
Comment in
- The need for speed.
Flicek P. Flicek P. Genome Biol. 2009;10(3):212. doi: 10.1186/gb-2009-10-3-212. Epub 2009 Mar 27. Genome Biol. 2009. PMID: 19344490 Free PMC article.
Similar articles
- Fast and accurate long-read alignment with Burrows-Wheeler transform.
Li H, Durbin R. Li H, et al. Bioinformatics. 2010 Mar 1;26(5):589-95. doi: 10.1093/bioinformatics/btp698. Epub 2010 Jan 15. Bioinformatics. 2010. PMID: 20080505 Free PMC article. - Fast and memory efficient approach for mapping NGS reads to a reference genome.
Kumar S, Agarwal S, Ranvijay. Kumar S, et al. J Bioinform Comput Biol. 2019 Apr;17(2):1950008. doi: 10.1142/S0219720019500082. J Bioinform Comput Biol. 2019. PMID: 31057068 - Ψ-RA: a parallel sparse index for genomic read alignment.
Oğuzhan Külekci M, Hon WK, Shah R, Scott Vitter J, Xu B. Oğuzhan Külekci M, et al. BMC Genomics. 2011;12 Suppl 2(Suppl 2):S7. doi: 10.1186/1471-2164-12-S2-S7. Epub 2011 Jul 27. BMC Genomics. 2011. PMID: 21989248 Free PMC article. - SRmapper: a fast and sensitive genome-hashing alignment tool.
Gontarz PM, Berger J, Wong CF. Gontarz PM, et al. Bioinformatics. 2013 Feb 1;29(3):316-21. doi: 10.1093/bioinformatics/bts712. Epub 2012 Dec 24. Bioinformatics. 2013. PMID: 23267171 - A survey of sequence alignment algorithms for next-generation sequencing.
Li H, Homer N. Li H, et al. Brief Bioinform. 2010 Sep;11(5):473-83. doi: 10.1093/bib/bbq015. Epub 2010 May 11. Brief Bioinform. 2010. PMID: 20460430 Free PMC article. Review.
Cited by
- Urogenital colonization and pathogenicity of E. Coli in the vaginal microbiota during pregnancy.
Boutouchent N, Vu TNA, Landraud L, Kennedy SP. Boutouchent N, et al. Sci Rep. 2024 Oct 26;14(1):25523. doi: 10.1038/s41598-024-76438-2. Sci Rep. 2024. PMID: 39462143 Free PMC article. - MicroRNA-mediated network redundancy is constrained by purifying selection and contributes to expression robustness in Drosophila melanogaster.
Dai A, Lan W, Lyu Y, Zhou X, Mi X, Tang T, Liufu Z. Dai A, et al. Commun Biol. 2024 Nov 4;7(1):1431. doi: 10.1038/s42003-024-07162-w. Commun Biol. 2024. PMID: 39496904 Free PMC article. - Whole-Transcriptome Analysis Reveals Potential CeRNA Regulatory Mechanism in Takifugu rubripes against Cryptocaryon irritans Infection.
Xia Y, Yu X, Yuan Z, Yang Y, Liu Y. Xia Y, et al. Biology (Basel). 2024 Oct 1;13(10):788. doi: 10.3390/biology13100788. Biology (Basel). 2024. PMID: 39452097 Free PMC article. - The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis.
Zheng F, Chen J, Zhang X, Wang Z, Chen J, Lin X, Huang H, Fu W, Liang J, Wu W, Li B, Yao H, Hu H, Song E. Zheng F, et al. Nat Commun. 2021 Feb 26;12(1):1341. doi: 10.1038/s41467-021-21535-3. Nat Commun. 2021. PMID: 33637716 Free PMC article. - Upstream distal regulatory elements contact the Lmo2 promoter in mouse erythroid cells.
Bhattacharya A, Chen CY, Ho S, Mitchell JA. Bhattacharya A, et al. PLoS One. 2012;7(12):e52880. doi: 10.1371/journal.pone.0052880. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285212 Free PMC article.
References
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. - DOI - PMC - PubMed
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. - DOI - PMC - PubMed
- Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J, Conyers J, Sander N, Shi X, Osborne JR, Minx P, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R01 GM083873/GM/NIGMS NIH HHS/United States
- R01-LM006845/LM/NLM NIH HHS/United States
- R01 LM006845-10/LM/NLM NIH HHS/United States
- R01 HG004885/HG/NHGRI NIH HHS/United States
- R01 LM006845-09/LM/NLM NIH HHS/United States
- R01 LM006845/LM/NLM NIH HHS/United States
- R01-GM083873/GM/NIGMS NIH HHS/United States
- R01 GM083873-06/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases