PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool - PubMed (original) (raw)
PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool
Wenjing Li et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Nuclear domains of promyelocytic leukemia protein (PML) are known to act as signaling nodes in many cellular processes. Although the impact of PML expression in driving cell fate decisions for injured cells is well established, the function of PML in the context of tissue development is less well understood. Here, the in vivo role of PML in developmental processes in the murine mammary gland has been investigated. Data are presented showing that expression of PML is tightly regulated by three members of the Stat family of transcription factors that orchestrate the functional development of the mammary secretory epithelium during pregnancy. Developmental phenotypes were also discovered in the virgin and pregnant Pml null mouse, typified by aberrant differentiation of mammary epithelia with reduced ductal and alveolar development. PML depletion was also found to disturb the balance of two distinct luminal progenitor populations. Overall, it is shown that PML is required for cell lineage determination in bi-potent luminal progenitor cells and that the precise regulation of PML expression is required for functional differentiation of alveolar cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
PML expression during mammary gland development. (A) Murine mammary glands from virgin (V), gestation (G), lactation (L), and involution (I) time points (in days) as indicated were probed for PML protein expression by immunoblotting. PML appears as two bands corresponding to the two alternatively spliced isoforms labeled 1 and 2 respectively. The expression levels of PML isoforms 1 and 2 relative to E-Cadherin (E-Cad) are shown in the lower panel. (B) Nuclear PML-NDs (red) were visualized by immunohistochemistry, with DAPI counterstain, on mammary gland sections from various stages of development. Scale bars, 5 μm. (C) RNA levels of PML isoforms 1 and 2 were measured by quantitative RT-PCR and are expressed relative to the level of cyclophilin mRNA. (D) KIM2 cells were differentiated with prolactin and dexamethasone for the number of days indicated and probed for PML expression. The transferrin receptor (TR) was used as a loading control.
Fig. 2.
Stat activity negatively regulates PML protein expression in vitro and in vivo. EpH4 cells were treated with (A) oncostatin M, (B) prolactin, or (C) IL-13 for the indicated times and were probed for active Stat 3, 5, and 6, respectively, and PML. A representative fluorescence micrograph of PML immunostaining after oncostatin M treatment is shown (A, lower panel), highlighting the reduction in number and intensity of PML-NDs after treatment. Scale bars, 5 μm. (D) Glands from wild-type (Stat3flox/flox) and Stat3 conditional knockout (BLG-Cre/Stat3flox/flox) mice at 24 hours' involution were analyzed for PML mRNA expression by quantitative RT-PCR and for protein expression by (E) immunofluorescence (scale bars, 5 μm) and (F) immunoblotting (left panel). Immunoblots were analyzed by densitometry (right panel) to indicate the increased level of PML expression in Stat3 null mice (n = 3, ***P < 0.01).
Fig. 3.
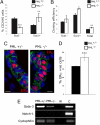
PML influences Stat activation and MGD. (A) Pml+/− and _Pml_−/− mammary glands from 15-day-gestation mice were analyzed for β-casein expression (red) by immunofluorescence (scale bars, 10 μm). (B) Glands from Pml+/− and _Pml_−/− mice at 15 days gestation were analyzed for Stat 5 and Stat 6 activation by immunoblotting.
Fig. 4.
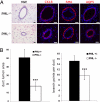
PML loss affects mammary gland morphogenesis. (A) Individual ducts from H&E-stained and immunostained sections of Pml+/− and _Pml_−/− mammary glands from 6-week-old virgin mice are shown, highlighting the smaller size but retained architecture of _Pml_−/− ducts. Immunostains are for cytokeratin 18 (CK18), smooth muscle actin (SMA), and aquaporin 5 (AQP5). (B) H&E-stained sections and whole mounts were quantified in terms of duct lumen cross-sectional area (n = 30, ***P < 0.01) and number of branch points per duct (n = 10, ***P < 0.01), respectively (lower panels).
Fig. 5.
PML loss influences mammary epithelial progenitor populations (A) Numbers of Sca1+ and Sca1− luminal progenitors shown as a percentage of live luminal cells (CD24hi) assayed by flow cytometry from 14-week-old Pml+/− and _Pml_−/− virgin mammary glands highlighting the reduction in Sca1− and increase in Sca1+ populations in KO tissue (n = 2, ***P < 0.01, **P < 0.02). (B) Cloning efficiency (per hundred cells) of FACS sorted Sca1+, Sca1− luminal progenitors and total epithelial cells from 14-week-old Pml+/− and _Pml_−/− mice (n = 2; ***P < 0.01, **P < 0.02). (C) Immunohistochemistry for ERα (red) and E-Cadherin (green) on mammary gland sections from 6-week-old Pml+/− and Pml−/− virgin mammary glands highlighting the increase in the number of ERα+ cells. (D) Counts of ERα+ cells in Pml+/− and _Pml_−/− virgin mammary glands (n = 3, ***P < 0.01). (E) Gata-3 and Notch-1 mRNA expression analyzed by RT-PCR from the sorted Sca1- cell populations of Pml+/− and _Pml_−/− glands. Control lanes are without RNA (labeled W) and RNA from a 15DG Pml+/− gland (labeled C). The absence of Notch-1 expression suggests that Stat6 activity is regulating Gata-3 expression.
References
- Salomoni P, Ferguson BJ, Wyllie AH, Rich T. New insights into the role of PML in tumour suppression. Cell Res. 2008;18:622–640. - PubMed
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. - PubMed
- Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2006NOVPHD11/BBC_/Breast Cancer Now/United Kingdom
- BB/D012937/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U132670601/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases