E. coli RNase III(E38A) generates discrete-sized products from long dsRNA - PubMed (original) (raw)
E. coli RNase III(E38A) generates discrete-sized products from long dsRNA
Jianping Xiao et al. RNA. 2009 May.
Abstract
Ribonuclease III (RNase III) represents a highly conserved family of double-strand-specific endoribonucleases that are important for RNA processing and post-transcriptional gene regulation in both prokaryotes and eukaryotes. We constructed a single amino acid substitution (E38A) of RNase III that shows a unique and useful enzymatic activity. It produces a dsRNA product of a discrete size migrating as 23 base pairs (bp) when given a long dsRNA as a substrate in an easy-to-control reaction. We demonstrate that the RNase III(E38A) mutant produces the 23-bp dsRNA product by making a double-strand cleavage of the long dsRNA substrate with the product being protected from further digestion. Using the hairpin RNA R1.1 as a substrate, RNase III(E38A) cleaves at the primary site and remains bound to the RNA, thereby preventing cleavage at the secondary site. The 23-bp dsRNA product is demonstrated to be a pool of dsRNAs representative of the long dsRNA substrate and has RNA interference activity in mammalian tissue culture transfection experiments. The RNA interference activity suggests that the 23-bp dsRNA product has typical 2-nucleotide 3' overhangs and behaves as siRNA thereby making it a useful tool in RNA interference experiments.
Figures
FIGURE 1.
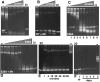
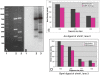
RNase activity for wild-type (WT) and E38A substitutions. A 900-bp dsRNA (500 ng) was cleaved with WT His-tagged RNase III or His-tagged E38A with a serial dilution of protein (illustrated by the gray triangle) using a starting amount of 4 μg. All reactions were performed as described in Materials and Methods. (A) WT RNase III. (B) WT RNase III with Mn2+ replacing Mg2+ in reaction buffer. (C) RNase III (E38A). (D) E38A with Mn2+ replacing Mg2+ in reaction buffer. (E) Reactions with E38A stopped at the times indicated in minutes. (F) Reactions with E38A stopped at the times indicated in days. “−” Indicates no protein in the reaction; “22” indicates a synthetic 22-nt dsRNA size marker. White arrow points to 23-bp dsRNA product; gray arrow points to smaller species product of WT RNase III.
FIGURE 2.
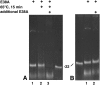
E38A product release requires denaturation. (A) A 900-bp dsRNA was incubated with E38A for 30 min at 37°C in reaction buffer, and one third was removed for electrophoresis (lane 1). The remaining two thirds was incubated for 15 min at 65°C, and one-half was removed for electrophoresis (lane 2). Additional E38A was added to the remaining third and incubated for 30 min at 37°C (lane 3). (B) A 900-bp dsRNA was incubated with E38A for 30 min at 37°C in reaction buffer, and one-half was removed for electrophoresis (lane 1). Additional E38A was added to the remaining half and incubated for 30 min at 37°C (lane 2). “22” Designates a synthetic 22-nte dsRNA size marker.
FIGURE 3.
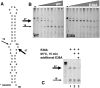
E38A digestion of hairpin RNA, R1.1. (A) R1.1 sequence and secondary structure are shown. Bold arrow indicates the primary cleavage site, and thin arrow indicates the secondary cleavage site. “*” indicates the fluorescein tag. (B) Fluorescein labeled R1.1 was incubated with increasing amounts of WT RNase III (left panel) or E38A (right panel) for 30 min at 37°C, electrophoresed on a denaturing polyacrylamide gel, and imaged with a PhosphorImager. Bold arrow indicates the product of cleavage at the primary site, and thin arrow indicates the product of cleavage at the secondary site. (C) Fluorescein labeled R1.1 was incubated with E38A for 30 min at 37°C in reaction buffer, and one-third was removed for electrophoresis (lane 1). The remaining two-thirds were incubated for 15 min at 65°C, and one-half was removed for electrophoresis (lane 2). Additional E38A was added to the remaining third and incubated for 30 min at 37°C (lane 3). “−” Indicates uncut R1.1.
FIGURE 4.
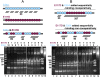
E38A and E117D combined digestion of dsRNA. E38A and E117D were used to digest a 900-bp dsRNA; total of 4 μg of the two proteins were incubated in the ratios indicated above the two gels (from 10:0 to 0:10). (Lane 1) dsRNA size markers. (Lanes 2_–_12) Digestions with E38A and E117D. (Lane 13) No enzyme. (A) Cartoon depicts mode of action of E38A alone (blue), E117D alone (maroon), and combined digestion when both proteins are added simultaneously to the reaction. Gel panel depicts results when both mutant enzymes are added to the substrate simultaneously at ratios indicated above the lanes. (B) Cartoon depicts mode of action for cooperative vs. noncooperative binding of RNase III to dsRNA when the two proteins are added sequentially, with E117D being added first. Top scenario shows the predicted results when binding is cooperative. Bottom scenario shows the predicted results when binding is noncooperative. Note that the predicted results for noncooperative binding are the same as if the two proteins are added simultaneously. The gel depicts the results when E117D is added first and E38A added 15 min later at ratios indicated above the lanes.
FIGURE 5.
dsRNA product generated with E38A represents a collection of 23-mers from its larger dsRNA substrate. (A) A 1112-bp fragment of dnmt1 was digested with AciI (lane 2) and BpmI (lane 3), electrophoresed on a 1.25% agarose gel, and imaged via EtBr staining. (Lane 1) Molecular weight size standards. (B) The gel was transferred to nitrocellulose and hybridized with 32P-labeled 23-bp dsRNA generated by E38A from a 1112-bp dsRNA transcribed from the same dnmt1 DNA fragment. Lanes 1_–_3 correspond to lanes 1–3 in A. (C) Bar graph of intensity (in pixels) of EtBr staining (purple) and hybridization intensity (gray) for each AciI-generated DNA fragment. (D) Bar graph of intensity (in pixels) of EtBr staining (purple) and hybridization intensity (gray) for each BpmI-generated DNA fragment.
FIGURE 6.
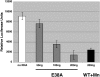
RNA interference activity of E38A-generated luciferase siRNA. NIH 3T3 cells were transfected with a plasmid expressing firefly luciferase and subsequently transfected with either E38A-generated luciferase siRNA or WT RNase III/Mn++-generated luciferase siRNA. Cells were harvested 48 h after the addition of siRNA and assayed for luciferase activity. Activities were normalized for transfection efficiency as described in Materials and Methods. No siRNA added, white bar; E38A-generated siRNA, gray bars; WT RNase III-generated siRNA, black bar.
Similar articles
- Coexpression of Escherichia coli RNase III in silkworm cells improves the efficiency of RNA interference induced by long hairpin dsRNAs.
Lee JM, Kojin Y, Tatsuke T, Mon H, Miyagawa Y, Kusakabe T. Lee JM, et al. Insect Sci. 2013 Feb;20(1):69-77. doi: 10.1111/j.1744-7917.2012.01569.x. Epub 2012 Nov 6. Insect Sci. 2013. PMID: 23955827 - Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain.
Sun W, Jun E, Nicholson AW. Sun W, et al. Biochemistry. 2001 Dec 11;40(49):14976-84. doi: 10.1021/bi011570u. Biochemistry. 2001. PMID: 11732918 - Ribonuclease III mechanisms of double-stranded RNA cleavage.
Nicholson AW. Nicholson AW. Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):31-48. doi: 10.1002/wrna.1195. Epub 2013 Sep 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24124076 Free PMC article. Review. - Structural basis for non-catalytic and catalytic activities of ribonuclease III.
Ji X. Ji X. Acta Crystallogr D Biol Crystallogr. 2006 Aug;62(Pt 8):933-40. doi: 10.1107/S090744490601153X. Epub 2006 Jul 18. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16855311 Review.
Cited by
- RNase III controls the degradation of corA mRNA in Escherichia coli.
Lim B, Sim SH, Sim M, Kim K, Jeon CO, Lee Y, Ha NC, Lee K. Lim B, et al. J Bacteriol. 2012 May;194(9):2214-20. doi: 10.1128/JB.00099-12. Epub 2012 Feb 17. J Bacteriol. 2012. PMID: 22343302 Free PMC article. - Expression of Human ACE2 N-terminal Domain, Part of the Receptor for SARS-CoV-2, in Fusion With Maltose-Binding Protein, E. coli Ribonuclease I and Human RNase A.
Xu SY, Fomenkov A, Chen TH, Yigit E. Xu SY, et al. Front Microbiol. 2021 Jun 11;12:660149. doi: 10.3389/fmicb.2021.660149. eCollection 2021. Front Microbiol. 2021. PMID: 34177838 Free PMC article. - Fitness and Functional Landscapes of the E. coli RNase III Gene rnc.
Weeks R, Ostermeier M. Weeks R, et al. Mol Biol Evol. 2023 Mar 4;40(3):msad047. doi: 10.1093/molbev/msad047. Mol Biol Evol. 2023. PMID: 36848192 Free PMC article. - Two tandem RNase III cleavage sites determine betT mRNA stability in response to osmotic stress in Escherichia coli.
Sim M, Lim B, Sim SH, Kim D, Jung E, Lee Y, Lee K. Sim M, et al. PLoS One. 2014 Jun 23;9(6):e100520. doi: 10.1371/journal.pone.0100520. eCollection 2014. PLoS One. 2014. PMID: 24956275 Free PMC article. - Pseudomonas mRNA 2.0: Boosting Gene Expression Through Enhanced mRNA Stability and Translational Efficiency.
Neves D, Vos S, Blank LM, Ebert BE. Neves D, et al. Front Bioeng Biotechnol. 2020 Jan 24;7:458. doi: 10.3389/fbioe.2019.00458. eCollection 2019. Front Bioeng Biotechnol. 2020. PMID: 32039175 Free PMC article.
References
- Blaszczyk J., Tropeae J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. - PubMed
- Court D.L. RNA processing and degradation by RNase III. In: Belasco J.G., Brawerman G., editors. Control of messenger RNA stability. Academic; New York: 1993. pp. 71–116.
- Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. - PubMed
- Jeltsch A. On the enzymatic properties of Dnmt1: Specificity, processivity, mechanism of linear diffusion, and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases