Metagenomic pyrosequencing and microbial identification - PubMed (original) (raw)
Review
Metagenomic pyrosequencing and microbial identification
Joseph F Petrosino et al. Clin Chem. 2009 May.
Abstract
Background: The Human Microbiome Project has ushered in a new era for human metagenomics and high-throughput next-generation sequencing strategies.
Content: This review describes evolving strategies in metagenomics, with a special emphasis on the core technology of DNA pyrosequencing. The challenges of microbial identification in the context of microbial populations are discussed. The development of next-generation pyrosequencing strategies and the technical hurdles confronting these methodologies are addressed. Bioinformatics-related topics include taxonomic systems, sequence databases, sequence-alignment tools, and classifiers. DNA sequencing based on 16S rRNA genes or entire genomes is summarized with respect to potential pyrosequencing applications.
Summary: Both the approach of 16S rDNA amplicon sequencing and the whole-genome sequencing approach may be useful for human metagenomics, and numerous bioinformatics tools are being deployed to tackle such vast amounts of microbiological sequence diversity. Metagenomics, or genetic studies of microbial communities, may ultimately contribute to a more comprehensive understanding of human health, disease susceptibilities, and the pathophysiology of infectious and immune-mediated diseases.
Figures
Figure 1. Pyrosequencing chemistry
This figure shows the biochemical reactions and enzymes involved in the generation of light signals by DNA pyrosequencing (18). Each peak in the pyrograms represents a pulse of light detected in the instrument. ATP, adenosine triphosphate; ADP, adenosine diphosphate; dNDP, deoxy-nucleotidyl diphosphate; dNMP, deoxy-nucleotidyl monophosphate; PPi, pyrophosphate. Adapted from
Figure 2. Conserved and hypervariable regions in the 16S rRNA gene
The interspersed conserved regions (C1–C9) are shown in gray, and the hypervariable regions (V1–V9) are depicted in different colors. An example of primer selection for DNA amplification and sequencing-based microbial identification is provided in the figure (V4 subregion with pink circles and arrows representing primer binding sites).
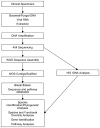
Figure 3. Deployment of 454 sequencing technology for metagenomics
A proposed pipeline is shown for high throughput 454 sequencing and associated bio-informatics strategies in metagenomics. Each box represents a discrete step in the process using either whole genome sequencing (WGS) or 16S rDNA amplicon sequencing. Note that WGS may be performed with prior whole genome amplification (WGA) or without prior amplification.
Similar articles
- Quantitative Assessment of Shotgun Metagenomics and 16S rDNA Amplicon Sequencing in the Study of Human Gut Microbiome.
Laudadio I, Fulci V, Palone F, Stronati L, Cucchiara S, Carissimi C. Laudadio I, et al. OMICS. 2018 Apr;22(4):248-254. doi: 10.1089/omi.2018.0013. OMICS. 2018. PMID: 29652573 - Microbial resolution of whole genome shotgun and 16S amplicon metagenomic sequencing using publicly available NEON data.
Brumfield KD, Huq A, Colwell RR, Olds JL, Leddy MB. Brumfield KD, et al. PLoS One. 2020 Feb 13;15(2):e0228899. doi: 10.1371/journal.pone.0228899. eCollection 2020. PLoS One. 2020. PMID: 32053657 Free PMC article. - A bioinformatics pipeline integrating predictive metagenomics profiling for the analysis of 16S rDNA/rRNA sequencing data originated from foods.
Mataragas M, Alessandria V, Ferrocino I, Rantsiou K, Cocolin L. Mataragas M, et al. Food Microbiol. 2018 Dec;76:279-286. doi: 10.1016/j.fm.2018.05.009. Epub 2018 May 24. Food Microbiol. 2018. PMID: 30166151 - Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.
Staley C, Sadowsky MJ. Staley C, et al. J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review. - Genomic sequencing of uncultured microorganisms from single cells.
Lasken RS. Lasken RS. Nat Rev Microbiol. 2012 Sep;10(9):631-40. doi: 10.1038/nrmicro2857. Nat Rev Microbiol. 2012. PMID: 22890147 Review.
Cited by
- A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria.
Chung HJ, Castro CM, Im H, Lee H, Weissleder R. Chung HJ, et al. Nat Nanotechnol. 2013 May;8(5):369-75. doi: 10.1038/nnano.2013.70. Epub 2013 May 5. Nat Nanotechnol. 2013. PMID: 23644570 Free PMC article. - Advancing quantitative PCR with color cycle multiplex amplification.
Chen W, Zhang K, Huang F, Zhao L, Waldren GC, Jiang Q, Chen SX, Wang B, Guo W, Zhang DY, Zhang JX. Chen W, et al. Nucleic Acids Res. 2024 Sep 23;52(17):e81. doi: 10.1093/nar/gkae683. Nucleic Acids Res. 2024. PMID: 39119904 Free PMC article. - Do you kiss your mother with that mouth? An authentic large-scale undergraduate research experience in mapping the human oral microbiome.
Wang JT, Daly JN, Willner DL, Patil J, Hall RA, Schembri MA, Tyson GW, Hugenholtz P. Wang JT, et al. J Microbiol Biol Educ. 2015 May 1;16(1):50-60. doi: 10.1128/jmbe.v16i1.816. eCollection 2015 May. J Microbiol Biol Educ. 2015. PMID: 25949757 Free PMC article. - Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis.
Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, Li C, Wei W, Liu J, Chen L, Davis C, Xu J. Huang S, et al. BMC Oral Health. 2011 Dec 12;11:33. doi: 10.1186/1472-6831-11-33. BMC Oral Health. 2011. PMID: 22152152 Free PMC article. - Isolation and characterization of a novel Sphingobium yanoikuyae strain variant that uses biohazardous saturated hydrocarbons and aromatic compounds as sole carbon sources.
Mitra M, Nguyen KM, Box TW, Gilpin JS, Hamby SR, Berry TL, Duckett EH. Mitra M, et al. F1000Res. 2020 Jul 24;9:767. doi: 10.12688/f1000research.25284.1. eCollection 2020. F1000Res. 2020. PMID: 32934808 Free PMC article.
References
- Wilson M. Bacteriology of Humans: An Ecological Perspective. Malden, MA: Blackwell Publishing; 2008. pp. 266–326.
- Thies FL, Konig W, Konig B. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J Med Microbiol. 2007;56:755–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 HG004973/HG/NHGRI NIH HHS/United States
- U54 HG003273-04S1/HG/NHGRI NIH HHS/United States
- P30 DK56338/DK/NIDDK NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R21AT003482/AT/NCCIH NIH HHS/United States
- R01 DK065075-01A2/DK/NIDDK NIH HHS/United States
- R21 AT003482/AT/NCCIH NIH HHS/United States
- R21 AT003482-01A1/AT/NCCIH NIH HHS/United States
- R01 AT004326-01A1/AT/NCCIH NIH HHS/United States
- R01 AT004326/AT/NCCIH NIH HHS/United States
- U54 HG004973-01/HG/NHGRI NIH HHS/United States
- R01 DK065075/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources