Functional organization of the S. cerevisiae phosphorylation network - PubMed (original) (raw)
. 2009 Mar 6;136(5):952-63.
doi: 10.1016/j.cell.2008.12.039.
Hannes Braberg, Monika Mehta, Gal Chechik, Gerard Cagney, Paromita Mukherjee, Andrea C Silva, Michael Shales, Sean R Collins, Sake van Wageningen, Patrick Kemmeren, Frank C P Holstege, Jonathan S Weissman, Michael-Christopher Keogh, Daphne Koller, Kevan M Shokat, Nevan J Krogan
Affiliations
- PMID: 19269370
- PMCID: PMC2856666
- DOI: 10.1016/j.cell.2008.12.039
Functional organization of the S. cerevisiae phosphorylation network
Dorothea Fiedler et al. Cell. 2009.
Abstract
Reversible protein phosphorylation is a signaling mechanism involved in all cellular processes. To create a systems view of the signaling apparatus in budding yeast, we generated an epistatic miniarray profile (E-MAP) comprised of 100,000 pairwise, quantitative genetic interactions, including virtually all protein and small-molecule kinases and phosphatases as well as key cellular regulators. Quantitative genetic interaction mapping reveals factors working in compensatory pathways (negative genetic interactions) or those operating in linear pathways (positive genetic interactions). We found an enrichment of positive genetic interactions between kinases, phosphatases, and their substrates. In addition, we assembled a higher-order map from sets of three genes that display strong interactions with one another: triplets enriched for functional connectivity. The resulting network view provides insights into signaling pathway regulation and reveals a link between the cell-cycle kinase, Cak1, the Fus3 MAP kinase, and a pathway that regulates chromatin integrity during transcription by RNA polymerase II.
Figures
Figure 1. Epistasis analysis of the yeast signaling machinery
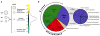
a) The entire spectrum of genetic interactions. Genetic interactions range from negative (e.g. synthetic sick) to positive (e.g. suppression) where the growth rate of the double mutant is either less ((aΔbΔ) < (aΔ)(bΔ)) or greater ((aΔbΔ) > (aΔ)(bΔ)) than the product of the growth rates of the corresponding single mutants, respectively. As shown in the distribution curve of our data set, significantly negative (S ≤ −2.5) and positive (S ≥ 2.0) genetic interactions are rare. b) Composition of the signaling E-MAP. For a full list of genes analyzed in this study, see Supplementary Table 1.
Figure 2. Comparison of the literature-derived signaling network to the genetic interaction data
a) A network diagram of characterized phosphorylation and dephosphorylation events between kinases and their substrates (green arrows) and phosphatases and their substrates (red arrows), manually curated from the literature (see Supplementary Table 2). Below this are specific examples of kinase-substrate (1) and phosphatase-substrate relationships (2), and cases where two kinases (3), two phosphatases (4) or one kinase and one phosphatase (5) target one or more substrates. Blue and yellow edges correspond to negative and positive genetic interactions, respectively. The thickness of the edge correlates with the strength of the genetic interaction. b) Ratios of highly positive (S ≥ 2.0) to negative (S ≤ −2.5) genetic interactions (P to N ratio). Above dashed line: The P to N ratios of previously published datasets (early secretory pathway (ESP)(Schuldiner et al., 2005) and chromosome function(Collins et al., 2007b)), the signaling E-MAP, only protein kinases and protein phosphatases (K vs. K or P vs. P), and protein kinases versus protein phosphatases (K vs. P) are presented. Below dashed line: The P to N ratios for known kinase-substrate and phosphatase-substrate pairs (K → S, P → S) (out of these 252 relationships that we genetically tested, we observed significant genetic interactions (positive and negative) for 25 of them (10%)), as well as kinase-kinase, phosphatase-phosphatase (K,K → S; P,P → S) and kinase-phosphatase pairs (K,P → S) that share one or more substrates are shown. To obtain these ratios and subsequent p-values, we also included regulatory subunits known to direct kinase/phosphatase activity towards specific substrates (see Methods and Supplementary Table 3).
Figure 3. Identification of factors involved in histone Htz1 depostion and acetylation
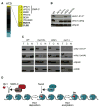
a) Full spectrum of HTZ1 genetic interactions. Genes with the strongest positive genetic interactions are highlighted. b) _clb2_Δ or _bud14_Δ profoundly reduce the total pool of Htz1-K14Ac. Whole cell extracts were isolated from isogenic strains containing Htz1 with a C-terminal HA3-tag, resolved by SDS-PAGE and immunoblotted as indicated. Rpn8 was used as a loading control. c) _bud14_Δ reduces both chromatin-associated Htz1 and Htz1-K14Ac, while _clb2_Δ specifically reduces the latter pool. Isogenic strains containing Htz1.HA3 were separated into T(otal), C(ytoplasmic) or N(uclear) fractions and immunoblotted with the indicated antibodies. Enrichment of the proteasome component Rpn8 and the chromatin component H2B in appropriate fractions (soluble cytoplasm and insoluble nucleus) demonstrate efficient separation. Using the H2B levels as a loading control, Htz1-K14Ac was decreased 3.7- and 3.0-fold in _bud14_Δ and _clb2_Δ respectively. By comparison, nuclear Htz1 was decreased 2.7-fold in _bud14_Δ but comparable to WT in clb2Δ. d) Schematic illustration of Htz1 deposition by SWR-C and acetylation by NuA4. The potential points of action of Bud14 and Clb2 in this pathway are indicated.
Figure 4. Mapping genetic interaction data onto known signaling pathways
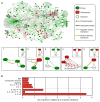
a) General schematic of the Sln1 branch of the HOG pathway. Protein kinases and phosphatases are denoted as green and red, respectively, while green and red arrows correspond to phosphorylation and dephosphorylation actions, respectively. b) The negative pathway regulators YPD1, PTC1, and NBP2, show strong positive genetic interactions with the pathway activators SSK1, SSK2, SSK22, PBS2, and HOG1. In contrast, genes coding for the protein phosphatases (PTC1, PTC2, PTC3, PTP2, and PTP3) acting on HOG1 show primarily negative genetic interactions among themselves. c) Scatter plot of the correlation coefficients of _pbs2_Δ and _hog1_Δ with all genetic profiles in the signaling E-MAP. d) Schematic illustration of the inositol polyphosphate pathway. e) A subset of genetic interactions for PLC1, KCS1, IPK1, and VIP1. In b) and e), negative and positive genetic interactions are indicated by blue or yellow squares, respectively. Gray squares represent genetic interactions not tested.
Figure 5. Triplet genetic motifs (TGMs) from the signaling E-MAP
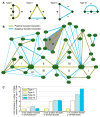
a) A schematic of the four types of triplet genetic motifs: all three positive (Type I); two positive, one negative (Type II); one positive, two negative (Type III); and all three negative (Type IV). b) A network diagram of all TGMs involving three kinases using highly negative (S ≤ −2.5) and positive (S ≥ 2.0) genetic interactions. The motifs were connected if they shared one or two nodes; blue and yellow edges correspond to negative and positive genetic interactions, respectively. The thickness of the edges is correlated with the strength of the genetic interaction. TGMs highlighted in grey are discussed in the text. c) Comparison of the ratio of the four types of TGMs that contain one or more kinase or phosphatase. The ratios are obtained by normalizing the number of genetic triplet motifs within each set (i.e. Types II, III, and IV) to the number of TGMs that are all positive (Type I). For a complete list of all TGMs, see Supplementary Table 5.
Figure 6. The Cak1 kinase functions in the Ctk1/Set2/Rpd3C(S) pathway that suppresses cryptic initiation by RNA polymerase II
a) Subset of Type I TGMs that share the Cak1-Set2 positive genetic interaction. For a complete picture of all Type I TGMs see Supplementary Figure 4. b) Tetrad analysis and serial 10-fold dilution spot testing demonstrates that the slow growth of the _cak1_-DAmP allele is strongly suppressed by _set2_Δ or _eaf3_Δ. c) CAK1 suppresses cryptic transcription initiation. The GAL1-FLO8-HIS3 reporter (Nourani et al., 2006) is described in Methods. The HIS3 gene product is only produced when transcription aberrantly initiates from a cryptic promoter within FLO8, as when chromatin structure in the transcribing gene is disrupted (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). His3 expression was monitored by 10-fold dilution spotting of the indicated strains onto synthetic complete (SC) medium +/− histidine with galactose/raffinose (2%/1% respectively) as the carbon source. The lower panels employed a series of _cak1_-temperature sensitive alleles (Espinoza et al., 1998) at their semi-permissive temperature (SC, 32°C, 48hrs; -HIS, 32°C, 96hrs). d) Possible schematic of this pathway controlling intergenic chromatin fidelity. e) The MAP kinase Fus3 regulates cryptic initiation. All strains contain a GAL1Sp-FLO8::HIS reporter, similar to that in Figure 6c (see Methods). Reporter strains were pinned in duplicate onto SC-HIS with galactose/raffinose as the carbon source, incubated at 30°C and photographed on days indicated. Panels are standardized to facilitate cross-comparison of reporter expression in each strain/time-point.
Similar articles
- Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map.
Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Collins SR, et al. Nature. 2007 Apr 12;446(7137):806-10. doi: 10.1038/nature05649. Epub 2007 Feb 21. Nature. 2007. PMID: 17314980 - Global control of histone modification by the anaphase-promoting complex.
Ramaswamy V, Williams JS, Robinson KM, Sopko RL, Schultz MC. Ramaswamy V, et al. Mol Cell Biol. 2003 Dec;23(24):9136-49. doi: 10.1128/MCB.23.24.9136-9149.2003. Mol Cell Biol. 2003. PMID: 14645525 Free PMC article. - Identification of putative negative regulators of yeast signaling through a screening for protein phosphatases acting on cell wall integrity and mating MAPK pathways.
Sacristán-Reviriego A, Martín H, Molina M. Sacristán-Reviriego A, et al. Fungal Genet Biol. 2015 Apr;77:1-11. doi: 10.1016/j.fgb.2015.02.011. Epub 2015 Feb 28. Fungal Genet Biol. 2015. PMID: 25736922 - The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae.
Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J. Roelants FM, et al. Biomolecules. 2017 Sep 5;7(3):66. doi: 10.3390/biom7030066. Biomolecules. 2017. PMID: 28872598 Free PMC article. Review. - Protein phosphatases of Saccharomyces cerevisiae.
Offley SR, Schmidt MC. Offley SR, et al. Curr Genet. 2019 Feb;65(1):41-55. doi: 10.1007/s00294-018-0884-y. Epub 2018 Sep 17. Curr Genet. 2019. PMID: 30225534 Free PMC article. Review.
Cited by
- Evolutionary and Biochemical Aspects of Chemical Stress Resistance in Saccharomyces cerevisiae.
Venancio TM, Bellieny-Rabelo D, Aravind L. Venancio TM, et al. Front Genet. 2012 Mar 30;3:47. doi: 10.3389/fgene.2012.00047. eCollection 2012. Front Genet. 2012. PMID: 22479268 Free PMC article. - Mathematical modelling of the MAP kinase pathway using proteomic datasets.
Tian T, Song J. Tian T, et al. PLoS One. 2012;7(8):e42230. doi: 10.1371/journal.pone.0042230. Epub 2012 Aug 8. PLoS One. 2012. PMID: 22905119 Free PMC article. - Srb5/Med18-mediated termination of transcription is dependent on gene looping.
Mukundan B, Ansari A. Mukundan B, et al. J Biol Chem. 2013 Apr 19;288(16):11384-94. doi: 10.1074/jbc.M112.446773. Epub 2013 Mar 8. J Biol Chem. 2013. PMID: 23476016 Free PMC article. - Functional phosphoproteomic mass spectrometry-based approaches.
López E, Wang X, Madero L, López-Pascual J, Latterich M. López E, et al. Clin Transl Med. 2012 Sep 5;1(1):20. doi: 10.1186/2001-1326-1-20. Clin Transl Med. 2012. PMID: 23369623 Free PMC article. - The Mck1 GSK-3 kinase inhibits the activity of Clb2-Cdk1 post-nuclear division.
McQueen J, van Dyk D, Young B, Loewen C, Measday V. McQueen J, et al. Cell Cycle. 2012 Sep 15;11(18):3421-32. doi: 10.4161/cc.21731. Epub 2012 Aug 23. Cell Cycle. 2012. PMID: 22918234 Free PMC article.
References
- Alcazar-Roman AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. - PubMed
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases