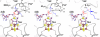Control of radical chemistry in the AdoMet radical enzymes - PubMed (original) (raw)
Review
Control of radical chemistry in the AdoMet radical enzymes
Kaitlin S Duschene et al. Curr Opin Chem Biol. 2009 Feb.
Abstract
The radical AdoMet superfamily comprises a diverse set of >2800 enzymes that utilize iron-sulfur clusters and S-adenosylmethionine (SAM or AdoMet) to initiate a diverse set of radical-mediated reactions. The intricate control these enzymes exercise over the radical transformations they catalyze is an amazing feat of elegance and sophistication in biochemistry. This review focuses on the accumulating evidence for how these enzymes control this remarkable chemistry, including controlling the reactivity between the iron-sulfur cluster and AdoMet, and controlling the subsequent radical transformations.
Figures
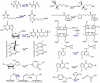
Figure 1
Representative reactions catalyzed by the radical-AdoMet enzymes. Abbreviations used: GRE-AE, glycyl radical activating enzymes such as pyruvate formate-lyase activating enzyme; LAM, lysine 2,3-aminomutase; BioB, biotin synthase; LipA, lipoyl synthase; MoaA, molybdopterin cofactor biosynthesis enzyme; HemN, oxygen-independent coproporphyrinogen oxidase; SPL, spore photoproduct lyase; MiaB, tRNA methylthiolation enzyme; ThiC and ThiH, enzymes involved in thiamine biosynthesis, TYW1, tRNA modification enzyme; AtsB, formylglycine-generating enzyme.
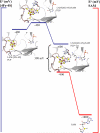
Figure 2
Common mechanistic steps proposed to be involved in the radical AdoMet enzymes.
Figure 3
Binding energetics and redox potentials in the radical AdoMet enzyme lysine 2,3-aminomutase. The blue scale indicates the changes in redox potential for the [4Fe-4S]2+/+ couple for the cluster in the free enzyme, for the cluster with the AdoMet bound, and for the cluster with AdoMet and lysine bound. The red scale indicates the redox potential for trialkylsulfoniums such as AdoMet (at -1800 mV) and for AdoMet bound to LAM and substrate (at -900 mV). The binding of AdoMet and substrate to the active site reduces the redox potential gap to ~300 mV.
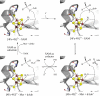
Figure 4
Views of the X-ray crystal structures of lysine 2,3-aminomutase (LAM, left, minus its C-terminus), MoaA (center), and pyruvate formate-lyase activating enzyme (PFL-AE, right), each with both AdoMet and substrate bound. Note the location of the radical-AdoMet cluster at one end of the TIM barrel, with AdoMet and substrate effectively sealing off the active site. Note also that as the substrate size increases from left to right, the size of the lateral opening in the TIM barrel also increases.
Figure 5
Views of the X-ray crystal structures of biotin synthase (BioB, left) and HydE (right), with the lid loop highlighted in orange.
Figure 6
Illustration of the results of ENDOR spectroscopic studies utilizing stabilized substrate and product radical analog intermediates. In all cases, van der Waals contacts are maintained between the 5'-methyl of 5'-deoxyadenosine (dAdo) and the substrate/product radicals. Illustrations are for the substrate radicals generated upon reaction with _trans_-4,5-dehydro-L-lysine (DHLys, left), 4-thia-L-lysine (SLys, center), and the product radical generated upon equilibration of the reduced state of the enzyme with AdoMet and L-α-lysine (right).
Comment in
- Frontiers in enzymatic C-H-bond activation.
Bollinger JM Jr, Broderick JB. Bollinger JM Jr, et al. Curr Opin Chem Biol. 2009 Feb;13(1):51-7. doi: 10.1016/j.cbpa.2009.03.018. Epub 2009 Apr 9. Curr Opin Chem Biol. 2009. PMID: 19362514 No abstract available.
Similar articles
- Radical AdoMet enzymes in complex metal cluster biosynthesis.
Duffus BR, Hamilton TL, Shepard EM, Boyd ES, Peters JW, Broderick JB. Duffus BR, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1254-63. doi: 10.1016/j.bbapap.2012.01.002. Epub 2012 Jan 14. Biochim Biophys Acta. 2012. PMID: 22269887 Review. - Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: a Mössbauer spectroscopic study.
Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. Krebs C, et al. J Am Chem Soc. 2002 Feb 13;124(6):912-3. doi: 10.1021/ja017562i. J Am Chem Soc. 2002. PMID: 11829592 - Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active 4Fe-4S cluster of pyruvate formate-lyase activating enzyme.
Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Walsby CJ, et al. J Am Chem Soc. 2002 Mar 27;124(12):3143-51. doi: 10.1021/ja012034s. J Am Chem Soc. 2002. PMID: 11902903 - Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.
Broderick WE, Hoffman BM, Broderick JB. Broderick WE, et al. Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review. - Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes.
Lanz ND, Booker SJ. Lanz ND, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1196-212. doi: 10.1016/j.bbapap.2012.07.009. Epub 2012 Jul 28. Biochim Biophys Acta. 2012. PMID: 22846545 Review.
Cited by
- Radiation inactivation of galactose oxidase, a monomeric enzyme with a stable free radical.
Kempner ES, Whittaker JW, Miller JH. Kempner ES, et al. Protein Sci. 2010 Feb;19(2):236-41. doi: 10.1002/pro.302. Protein Sci. 2010. PMID: 19998406 Free PMC article. - Complete stereospecific repair of a synthetic dinucleotide spore photoproduct by spore photoproduct lyase.
Silver SC, Chandra T, Zilinskas E, Ghose S, Broderick WE, Broderick JB. Silver SC, et al. J Biol Inorg Chem. 2010 Aug;15(6):943-55. doi: 10.1007/s00775-010-0656-8. Epub 2010 Apr 20. J Biol Inorg Chem. 2010. PMID: 20405152 Free PMC article. - Radical mediated ring formation in the biosynthesis of the hypermodified tRNA base wybutosine.
Young AP, Bandarian V. Young AP, et al. Curr Opin Chem Biol. 2013 Aug;17(4):613-8. doi: 10.1016/j.cbpa.2013.05.035. Epub 2013 Jul 12. Curr Opin Chem Biol. 2013. PMID: 23856057 Free PMC article. Review. - Iron-sulfur world in aerobic and hyperthermoacidophilic archaea Sulfolobus.
Iwasaki T. Iwasaki T. Archaea. 2010 Sep 19;2010:842639. doi: 10.1155/2010/842639. Archaea. 2010. PMID: 20885930 Free PMC article. Review. - Radical SAM enzymes: surprises along the path to understanding mechanism.
Broderick WE, Broderick JB. Broderick WE, et al. J Biol Inorg Chem. 2019 Sep;24(6):769-776. doi: 10.1007/s00775-019-01706-w. Epub 2019 Sep 7. J Biol Inorg Chem. 2019. PMID: 31494759 Free PMC article. Review.
References
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. - PMC - PubMed
- Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. - PubMed
- ** This is the most current general review on the radical SAM superfamily, which discusses the biochemical activities and crystal structures for the radical SAM enzymes that have been characterized to date. This review presents the enzymes according to their reactivity with SAM, either as a cofactor or substrate.
- Walsby C, Ortillo D, Yang J, Nnyepi M, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “Radical SAM” protein superfamily. Inorg. Chem. 2005;44:727–741. - PubMed
- Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S]+ cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 2002;124:3143–3151. - PubMed
- Walsby CJ, Ortillo D, Broderick WE, Broderick JB, Hoffman BM. An anchoring role for FeS Clusters: Chelation of the amino acid moiety of S-adenosylmethionine to the unique iron site of the [4Fe-4S] cluster of pyruvate formate-lyase activating enzyme. J. Am. Chem. Soc. 2002;124:11270–11271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources