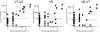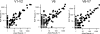Effects of experimental choices and analysis noise on surveys of the "rare biosphere" - PubMed (original) (raw)
Comparative Study
. 2009 May;75(10):3263-70.
doi: 10.1128/AEM.01931-08. Epub 2009 Mar 6.
Affiliations
- PMID: 19270149
- PMCID: PMC2681628
- DOI: 10.1128/AEM.01931-08
Comparative Study
Effects of experimental choices and analysis noise on surveys of the "rare biosphere"
Timothy J Hamp et al. Appl Environ Microbiol. 2009 May.
Abstract
When planning a survey of 16S rRNA genes from a complex environment, investigators face many choices including which primers to use and how to taxonomically classify sequences. In this study, we explored how these choices affected a survey of microbial diversity in a sample taken from the aerobic basin of the activated sludge of a North Carolina wastewater treatment plant. We performed pyrosequencing reactions on PCR products generated from primers targeting the V1-V2, V6, and V6-V7 variable regions of the 16S rRNA gene. We compared these sequences to 16S rRNA gene sequences found in a whole-genome shotgun pyrosequencing run performed on the same sample. We found that sequences generated from primers targeting the V1-V2 variable region had the best match to the whole-genome shotgun reaction across a range of taxonomic classifications from phylum to family. Pronounced differences between primer sets, however, occurred in the "rare biosphere" involving taxa that we observed in fewer than 11 sequences. We also examined the results of analysis strategies comparing a classification scheme using a nearest-neighbor approach to directly classifying sequences with a naïve Bayesian algorithm. Again, we observed pronounced differences between these analysis schemes in infrequently observed taxa. We conclude that if a study is meant to probe the rare biosphere, both the experimental conditions and analysis choices will have a profound impact on the observed results.
Figures
FIG. 1.
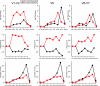
Sequence conservation as a function of alignment position for the 489,840 sequences in version 9.59 of the RDP. The x axis shows the position in the alignment as numbered by the E. coli 16S rRNA gene. The y axis shows the Shannon sequence entropy (see Materials and Methods), a widely used measure of conservation in multiple sequence alignments (23). Highly conserved positions within the alignment have a sequence entropy close to zero and hence are shown toward the top of the y axis. Positions of the hypervariable regions V1-V3 and V6-V7 are derived from Chakravorty et al. (3).
FIG. 2.
Number of sequences assigned at the family classification level by the RDP classification algorithm to different sequences for the V1-V2, V6, and V6-V7 primers plotted against the number of 16S sequences assigned to the whole-genome shotgun sequence set. One has been added to each count to allow the data to be shown on a log-log plot.
FIG. 3.
Across taxonomic levels, the results of a linear regression on log-transformed data between sequences generated by PCR targeting the 16S rRNA gene and 16S sequences culled from our whole-genome shotgun sequence set. Assignments are by the RDP classification algorithm as in Fig. 2. For each sequence set, two separate regressions were constructed, one for the rare biosphere (circles) with taxa seen 10 or fewer times in that sequence set's PCRs and one for a common biosphere (squares) with taxa seen 11 or more times in the PCR reactions (Fig. 1, gray lines). The top panels show the −log10 of the P value of the null hypothesis that the slope of the regression equals zero. The middle panels show Pearson's R values while the bottom panel shows the number of taxa for which classifications are made. Note that a significant P value (top panel) can be produced by either a negative or positive correlation.
FIG. 4.
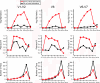
Comparison of classifications at the family level made by JGast and the RDP classification algorithm.
FIG. 5.
Regressions across classification levels on log-transformed data showing the comparison between the RDP classification algorithm and JGast. Two regressions were constructed for each comparison: one for a common biosphere in which a taxon was observed 11 or more times under either JGast or RDP (squares) and one for a rare taxon in which fewer than 11 taxa were observed under both classification schemes.
Similar articles
- Group-specific PCR primers for the phylum Acidobacteria designed based on the comparative analysis of 16S rRNA gene sequences.
Lee SH, Cho JC. Lee SH, et al. J Microbiol Methods. 2011 Aug;86(2):195-203. doi: 10.1016/j.mimet.2011.05.003. Epub 2011 May 12. J Microbiol Methods. 2011. PMID: 21600936 - A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments.
Li M, Hong Y, Klotz MG, Gu JD. Li M, et al. Appl Microbiol Biotechnol. 2010 Mar;86(2):781-90. doi: 10.1007/s00253-009-2361-5. Epub 2010 Jan 27. Appl Microbiol Biotechnol. 2010. PMID: 20107988 - Scratching the surface of the rare biosphere with ribosomal sequence tag primers.
Neufeld JD, Li J, Mohn WW. Neufeld JD, et al. FEMS Microbiol Lett. 2008 Jun;283(2):146-53. doi: 10.1111/j.1574-6968.2008.01124.x. Epub 2008 Apr 21. FEMS Microbiol Lett. 2008. PMID: 18429998 - Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys.
Youssef N, Sheik CS, Krumholz LR, Najar FZ, Roe BA, Elshahed MS. Youssef N, et al. Appl Environ Microbiol. 2009 Aug;75(16):5227-36. doi: 10.1128/AEM.00592-09. Epub 2009 Jun 26. Appl Environ Microbiol. 2009. PMID: 19561178 Free PMC article. - Review and re-analysis of domain-specific 16S primers.
Baker GC, Smith JJ, Cowan DA. Baker GC, et al. J Microbiol Methods. 2003 Dec;55(3):541-55. doi: 10.1016/j.mimet.2003.08.009. J Microbiol Methods. 2003. PMID: 14607398 Review.
Cited by
- Considerations For Optimizing Microbiome Analysis Using a Marker Gene.
de la Cuesta-Zuluaga J, Escobar JS. de la Cuesta-Zuluaga J, et al. Front Nutr. 2016 Aug 8;3:26. doi: 10.3389/fnut.2016.00026. eCollection 2016. Front Nutr. 2016. PMID: 27551678 Free PMC article. Review. - Assessment of bacterial endosymbiont diversity in Otiorhynchus spp. (Coleoptera: Curculionidae) larvae using a multitag 454 pyrosequencing approach.
Hirsch J, Strohmeier S, Pfannkuchen M, Reineke A. Hirsch J, et al. BMC Microbiol. 2012 Jan 18;12 Suppl 1(Suppl 1):S6. doi: 10.1186/1471-2180-12-S1-S6. BMC Microbiol. 2012. PMID: 22376125 Free PMC article. - Exploring biogeographic patterns of bacterioplankton communities across global estuaries.
Ghosh A, Bhadury P. Ghosh A, et al. Microbiologyopen. 2019 May;8(5):e00741. doi: 10.1002/mbo3.741. Epub 2018 Oct 10. Microbiologyopen. 2019. PMID: 30303297 Free PMC article. - Long-Term Survey Is Necessary to Reveal Various Shifts of Microbial Composition in Corals.
Yang SH, Tseng CH, Huang CR, Chen CP, Tandon K, Lee STM, Chiang PW, Shiu JH, Chen CA, Tang SL. Yang SH, et al. Front Microbiol. 2017 Jun 13;8:1094. doi: 10.3389/fmicb.2017.01094. eCollection 2017. Front Microbiol. 2017. PMID: 28659905 Free PMC article. - The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities.
Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC. Lee CK, et al. ISME J. 2012 May;6(5):1046-57. doi: 10.1038/ismej.2011.170. Epub 2011 Dec 15. ISME J. 2012. PMID: 22170424 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources