Tumor suppressors and cell metabolism: a recipe for cancer growth - PubMed (original) (raw)
Review
Tumor suppressors and cell metabolism: a recipe for cancer growth
Russell G Jones et al. Genes Dev. 2009.
Abstract
Growing tumors face two major metabolic challenges-how to meet the bioenergetic and biosynthetic demands of increased cell proliferation, and how to survive environmental fluctuations in external nutrient and oxygen availability when tumor growth outpaces the delivery capabilities of the existing vasculature. Cancer cells display dramatically altered metabolic circuitry that appears to directly result from the oncogenic mutations selected during the tumorigenic process. An emerging theme in cancer biology is that many of the genes that can initiate tumorigenesis are intricately linked to metabolic regulation. In turn, it appears that a number of well-established tumor suppressors play critical roles in suppressing growth and/or proliferation when intracellular supplies of essential metabolites become reduced. In this review, we consider the potential role of tumor suppressors as metabolic regulators.
Figures
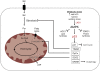
Figure 1.
Metabolic pathways of growth and proliferation. Growth factor-independent activation of the PI3K/Akt and c-Myc pathways drives changes in cellular metabolism to promote cancer cell growth and proliferation. PI3K/Akt and c-Myc facilitate increased rates of glucose uptake and glycolysis. Lactate dehydrogenase (LDH-A) maintains glycolytic flux by converting excess pyruvate to lactate and regenerating NAD+ in the process. Akt signaling promotes increased surface expression of the glucose transporter Glut1 and enhances the activity of glycolytic enzymes. Glucose (Glc) has multiple catabolic and biosynthetic fates, including glycolytic processing for production of ATP and pyruvate, processing through the pentose phosphate shunt to generate ribose 5-phosphate (Rib-5-P) and NADPH for nucleotide biosynthesis, or entry into the mitochondrion for biosynthesis or ATP generation by the TCA cycle and electron transport chain (ETC). Glucose-derived citrate (Cit) is exported to the cytosol and processed by ATP citrate lyase (ACL) to acetyl-CoA (Ac-CoA), which is channeled into lipid production. Glutamine (Gln) is deaminated to form glutamate, which can be processed further in the mitochondria by glutamate dehydrogenase (GDH) to generate α-ketoglutarate (αKG) and maintain TCA cycle function. Reactive oxygen species (ROS) are the result of mitochondrial OXPHOS. The TORC1 complex, which contains mTOR and its binding partner Raptor, regulates protein translation. mTOR coordinates surface expression of amino acid (AA) transporters, and promotes translation initiation by stimulating ribosomal S6 kinase activity and relieving 4E-BP-mediated inhibition of eIF4E. Amino acids stimulate mTOR activity through the activity of the Rag GTPases. PI3K/Akt signals activate mTOR primarily through inhibition of the TSC1–TSC2 complex, thus releasing inhibition of RhebGTP and activating mTOR. Bioenergetic signals antagonize the TORC1 complex through the LKB1–AMPK pathway. Known oncogenes are shown in green, tumor suppressors in red, and effectors with undefined roles in tumorigenesis are shown in blue.
Figure 2.
Metabolic stress during tumor development. A solid tumor can outstrip its nutrient and oxygen supply as it grows, resulting in metabolic stress (tumors experiencing metabolic stress are depicted in gray). As a consequence, tumor cells must undergo a period of metabolic adaptation to survive this metabolic stress or undergo apoptosis. Angiogenesis and neovascularization of the tumor is one strategy of metabolic adaptation used by tumors to relieve this stress.
Figure 3.
Strategies of metabolic adaptation in cancer. Intracellular sensors of energy, nutrients, and oxygen promote metabolic adaptation to stress during tumorigenesis. Major physiological strategies of metabolic adaptation include cell cycle inhibition, inhibition of biosynthetic pathways (lipid, protein synthesis), increases in bioenergetic pathways (β-oxidation, glycolysis, and OXPHOS), and induction of autophagy. Bioenergetic stress activates AMPK through an LKB-dependent manner. AMPK mediates metabolic adaptation through several pathways, including down-regulation of fatty acid synthesis through inhibition of ACC1, translation inhibition by blocking TORC1 activity, and activation of p53. Rag GTPases regulate TORC1 activity and autophagy in response to amino acid deprivation. Low HIF1α protein levels are maintained in cells under normoxia through the action of VHL-mediated proteosomal degradation. Low O2, ROS, or accumulation of the TCA substrates succinate or fumarate promotes HIF1α protein stabilization and transcriptional activity. HIF1α blocks pyruvate entry into the mitochondrion by up-regulating the PDH antagonist PDK1. Beclin-1 is required for autophagy induction and is antagonized by the anti-apoptotic proteins Bcl2/BclXL. Failure to manage or correct bioenergetic imbalance leads to cytochrome c release from mitochondria and induction of apoptosis. Oncogenes are displayed in green and tumor suppressors in red.
Figure 4.
The AMPK–p53 pathway in metabolic adaptation. Activation of the tumor suppressor p53 by AMPK promotes physiological adaptation to metabolic stress by multiple mechanisms. AMPK-dependent p53 activation negatively regulates cell cycle progression via the cyclin-dependent kinase (cdk) inhibitor p21. Impairment of glycolytic flux by TIGAR, combined with increased activation of β-oxidation and electron transport through expression of carnitine palmitoyltransferase Ic (Cpt1c) and SCO2, respectively, results in a p53-dependent metabolic shift toward OXPHOS. Transcriptional regulation of DRAM by p53 induces autophagy, the products of which can also fuel OXPHOS.
Similar articles
- Autophagy signaling and the cogwheels of cancer.
Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Botti J, et al. Autophagy. 2006 Apr-Jun;2(2):67-73. doi: 10.4161/auto.2.2.2458. Epub 2006 Apr 29. Autophagy. 2006. PMID: 16874041 Review. - Therapeutic targets in the ARF tumor suppressor pathway.
Saporita AJ, Maggi LB Jr, Apicelli AJ, Weber JD. Saporita AJ, et al. Curr Med Chem. 2007;14(17):1815-27. doi: 10.2174/092986707781058869. Curr Med Chem. 2007. PMID: 17627519 Free PMC article. Review. - Links between metabolism and cancer.
Dang CV. Dang CV. Genes Dev. 2012 May 1;26(9):877-90. doi: 10.1101/gad.189365.112. Genes Dev. 2012. PMID: 22549953 Free PMC article. Review. - Cancer as a metabolic disease and diabetes as a cancer risk?
Kankova K, Hrstka R. Kankova K, et al. Klin Onkol. 2012;25 Suppl 2:2S26-31. Klin Onkol. 2012. PMID: 23581013 Review. - Fueling immunity: insights into metabolism and lymphocyte function.
Pearce EL, Poffenberger MC, Chang CH, Jones RG. Pearce EL, et al. Science. 2013 Oct 11;342(6155):1242454. doi: 10.1126/science.1242454. Science. 2013. PMID: 24115444 Free PMC article. Review.
Cited by
- Spatial reorganization of Saccharomyces cerevisiae enolase to alter carbon metabolism under hypoxia.
Miura N, Shinohara M, Tatsukami Y, Sato Y, Morisaka H, Kuroda K, Ueda M. Miura N, et al. Eukaryot Cell. 2013 Aug;12(8):1106-19. doi: 10.1128/EC.00093-13. Epub 2013 Jun 7. Eukaryot Cell. 2013. PMID: 23748432 Free PMC article. - Hetastarch induced volume changes of starving adenoma and suggested mechanism of action.
Jakabčin P, Kello M, Záň J, Kolář J, Uličný J. Jakabčin P, et al. Sci Rep. 2022 Oct 26;12(1):17988. doi: 10.1038/s41598-022-21693-4. Sci Rep. 2022. PMID: 36289259 Free PMC article. - The role of AMPK in metabolism and its influence on DNA damage repair.
Szewczuk M, Boguszewska K, Kaźmierczak-Barańska J, Karwowski BT. Szewczuk M, et al. Mol Biol Rep. 2020 Nov;47(11):9075-9086. doi: 10.1007/s11033-020-05900-x. Epub 2020 Oct 18. Mol Biol Rep. 2020. PMID: 33070285 Free PMC article. Review. - Introduction to the molecular basis of cancer metabolism and the Warburg effect.
Ngo DC, Ververis K, Tortorella SM, Karagiannis TC. Ngo DC, et al. Mol Biol Rep. 2015 Apr;42(4):819-23. doi: 10.1007/s11033-015-3857-y. Mol Biol Rep. 2015. PMID: 25672512 Review. - Inhibition of Aerobic Glycolysis Represses Akt/mTOR/HIF-1α Axis and Restores Tamoxifen Sensitivity in Antiestrogen-Resistant Breast Cancer Cells.
Woo YM, Shin Y, Lee EJ, Lee S, Jeong SH, Kong HK, Park EY, Kim HK, Han J, Chang M, Park JH. Woo YM, et al. PLoS One. 2015 Jul 9;10(7):e0132285. doi: 10.1371/journal.pone.0132285. eCollection 2015. PLoS One. 2015. PMID: 26158266 Free PMC article.
References
- Aita V.M., Liang X.H., Murty V.V., Pincus D.L., Yu W., Cayanis E., Kalachikov S., Gilliam T.C., Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. - PubMed
- Bardeesy N., Sinha M., Hezel A.F., Signoretti S., Hathaway N.A., Sharpless N.E., Loda M., Carrasco D.R., DePinho R.A. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. - PubMed
- Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources