Finding the fifth base: genome-wide sequencing of cytosine methylation - PubMed (original) (raw)
Review
Finding the fifth base: genome-wide sequencing of cytosine methylation
Ryan Lister et al. Genome Res. 2009 Jun.
Abstract
Complete sequences of myriad eukaryotic genomes, including several human genomes, are now available, and recent dramatic developments in DNA sequencing technology are opening the floodgates to vast volumes of sequence data. Yet, despite knowing for several decades that a significant proportion of cytosines in the genomes of plants and animals are present in the form of methylcytosine, until very recently the precise locations of these modified bases have never been accurately mapped throughout a eukaryotic genome. Advanced "next-generation" DNA sequencing technologies are now enabling the global mapping of this epigenetic modification at single-base resolution, providing new insights into the regulation and dynamics of DNA methylation in genomes.
Figures
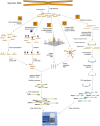
Figure 1.
Techniques for genome-wide sequencing of cytosine methylation sites. Three techniques used recently to generate bisulfite (BS) sequencing libraries compatible with next-generation sequencing are depicted. (A) MethylC-seq (Lister et al. 2008). Double-stranded universal adapter sequences in which all cytosines are methylated are ligated to fragmented genomic DNA. Sodium bisulfite treatment converts unmethylated cytosines to thymine, after which library yield enrichment by PCR with primers complementary to the universal adapter sequences produces the final library that can be sequenced. (B) BS-seq (Cokus et al. 2008). Ligation of a first set of double-stranded adaptors that contained methylated adenine bases within DpnI restriction sites close to the site of ligation with genomic DNA. After BS conversion, PCR is performed using primers complementary to the converted adapter sequences, yielding double-stranded DNA that is digested with DpnI to remove only the first adapter set. Sequencing adapters are subsequently ligated to the double-stranded BS-converted genomic DNA fragments, and PCR with primers complementary to the adapters performed to yield a sequencing library. (C) Reduced representation BS sequencing (RRBS) (Meissner et al. 2008). Genomic DNA is first digested by the methylation-insensitive MspI restriction enzyme, which cleaves the phosphodiester bond upstream of the CpG dinuclotide in its CCGG recognition element. Digested DNA is then separated by gel electrophoresis, and one or more specific size fractions are selected. The size-selected DNA is then end repaired, ligated to double-stranded methylated sequencing adapters (as described above for MethylC-seq), BS converted, and amplified by PCR with primers complementary to the adapter sequences.
Figure 2.
Techniques for enrichment of methylated or target regions prior to BS sequencing. Five approaches that may be used to reduce the complexity of a sample before BS conversion and next-generation sequencing are depicted, targeting methylated regions or select target sequences. (A) MeDIP. Methylated fragments of genomic DNA are immunoprecipitated with an anti-5-methylcytosine antibody. Purified, immunoprecipitated DNA is ligated to double-stranded universal adapter sequences in which all cytosines are methylated. Sodium bisulfite treatment converts unmethylated cytosines to thymine, after which library yield enrichment by PCR with primers complementary to the universal adapter sequences produces the final library that can be sequenced. (B) MBD. Methylated fragments of genomic DNA are isolated from a complex mix of fragmented genomic DNA with a methyl binding domain protein, after which adapter ligation, BS conversion, and PCR enrichment are performed as in A. (C) Microarray capture. Target sequences within a complex mix of fragmented genomic DNA are captured by hybridization to specific oligonucleotides on the surface of a microarray. Following isolation of the hybridized genomic DNA, adapter ligation, BS conversion, and PCR enrichment are performed as in A. (D) Capture in solution. Specific target regions within a mix of fragmented genomic DNA are captured by hybridization to specific oligonucleotides attached to beads in solution. Following isolation of the hybridized genomic DNA, adapter ligation, BS conversion, and PCR enrichment are performed as in A. (E) Molecular inversion probe capture. Fragmented genomic DNA is BS converted, after which molecular inversion probes are added that are designed to hybridize to specific target sequences after conversion. Polymerization primed by the 3′ end of the molecular inversion probe followed by ligation generates a circular molecule that contains the target sequence and is not digested by subsequent exonuclease treatment. PCR using primers that hybridize to the ends of the molecular inversion probes allows amplification of the target region, to which double-stranded universal adapter sequences are ligated to produce a library that is sufficient for next-generation sequencing.
Similar articles
- Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning.
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Cokus SJ, et al. Nature. 2008 Mar 13;452(7184):215-9. doi: 10.1038/nature06745. Epub 2008 Feb 17. Nature. 2008. PMID: 18278030 Free PMC article. - Post-conversion targeted capture of modified cytosines in mammalian and plant genomes.
Li Q, Suzuki M, Wendt J, Patterson N, Eichten SR, Hermanson PJ, Green D, Jeddeloh J, Richmond T, Rosenbaum H, Burgess D, Springer NM, Greally JM. Li Q, et al. Nucleic Acids Res. 2015 Jul 13;43(12):e81. doi: 10.1093/nar/gkv244. Epub 2015 Mar 26. Nucleic Acids Res. 2015. PMID: 25813045 Free PMC article. - Bisulfite methylation profiling of large genomes.
Reinders J, Paszkowski J. Reinders J, et al. Epigenomics. 2010 Apr;2(2):209-20. doi: 10.2217/epi.10.6. Epigenomics. 2010. PMID: 22121871 Review. - Whole-Genome Bisulfite Sequencing Protocol for the Analysis of Genome-Wide DNA Methylation and Hydroxymethylation Patterns at Single-Nucleotide Resolution.
Derbala D, Garnier A, Bonnet E, Deleuze JF, Tost J. Derbala D, et al. Methods Mol Biol. 2024;2842:353-382. doi: 10.1007/978-1-0716-4051-7_18. Methods Mol Biol. 2024. PMID: 39012605 - Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond.
Lister R, Gregory BD, Ecker JR. Lister R, et al. Curr Opin Plant Biol. 2009 Apr;12(2):107-18. doi: 10.1016/j.pbi.2008.11.004. Epub 2009 Jan 20. Curr Opin Plant Biol. 2009. PMID: 19157957 Free PMC article. Review.
Cited by
- Comprehensive DNA methylation analysis of the Aedes aegypti genome.
Falckenhayn C, Carneiro VC, de Mendonça Amarante A, Schmid K, Hanna K, Kang S, Helm M, Dimopoulos G, Fantappié MR, Lyko F. Falckenhayn C, et al. Sci Rep. 2016 Nov 2;6:36444. doi: 10.1038/srep36444. Sci Rep. 2016. PMID: 27805064 Free PMC article. - Analyses of Methylomes Derived from Meso-American Common Bean (Phaseolus vulgaris L.) Using MeDIP-Seq and Whole Genome Sodium Bisulfite-Sequencing.
Crampton M, Sripathi VR, Hossain K, Kalavacharla V. Crampton M, et al. Front Plant Sci. 2016 Apr 26;7:447. doi: 10.3389/fpls.2016.00447. eCollection 2016. Front Plant Sci. 2016. PMID: 27199997 Free PMC article. - Anchor-based bisulfite sequencing determines genome-wide DNA methylation.
Chapin N, Fernandez J, Poole J, Delatte B. Chapin N, et al. Commun Biol. 2022 Jun 16;5(1):596. doi: 10.1038/s42003-022-03543-1. Commun Biol. 2022. PMID: 35710818 Free PMC article. - DNA methylation estimation using methylation-sensitive restriction enzyme bisulfite sequencing (MREBS).
Bonora G, Rubbi L, Morselli M, Ma F, Chronis C, Plath K, Pellegrini M. Bonora G, et al. PLoS One. 2019 Apr 4;14(4):e0214368. doi: 10.1371/journal.pone.0214368. eCollection 2019. PLoS One. 2019. PMID: 30946758 Free PMC article. - Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2.
Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Kinde B, et al. Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6800-6. doi: 10.1073/pnas.1411269112. Epub 2015 Mar 4. Proc Natl Acad Sci U S A. 2015. PMID: 25739960 Free PMC article. Review.
References
- Absalan F., Ronaghi M. Molecular inversion probe assay. Methods Mol. Biol. 2007;396:315–330. - PubMed
- Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. - PubMed
- Bashiardes S., Veile R., Helms C., Mardis E.R., Bowcock A.M., Lovett M. Direct genomic selection. Nat. Methods. 2005;2:63–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources