CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation - PubMed (original) (raw)
. 2009 Mar 16;206(3):595-606.
doi: 10.1084/jem.20081385. Epub 2009 Mar 9.
Darin K Fogg, Emilie Narni-Mancinelli, Brigitte Senechal, Celine Trouillet, Noah Saederup, Julia Leemput, Karine Bigot, Laura Campisi, Marc Abitbol, Thierry Molina, Israel Charo, David A Hume, Ana Cumano, Gregoire Lauvau, Frederic Geissmann
Affiliations
- PMID: 19273628
- PMCID: PMC2699130
- DOI: 10.1084/jem.20081385
CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation
Cedric Auffray et al. J Exp Med. 2009.
Abstract
CX(3)CR1 expression is associated with the commitment of CSF-1R(+) myeloid precursors to the macrophage/dendritic cell (DC) lineage. However, the relationship of the CSF-1R(+) CX(3)CR1(+) macrophage/DC precursor (MDP) with other DC precursors and the role of CX(3)CR1 in macrophage and DC development remain unclear. We show that MDPs give rise to conventional DCs (cDCs), plasmacytoid DCs (PDCs), and monocytes, including Gr1(+) inflammatory monocytes that differentiate into TipDCs during infection. CX(3)CR1 deficiency selectively impairs the recruitment of blood Gr1(+) monocytes in the spleen after transfer and during acute Listeria monocytogenes infection but does not affect the development of monocytes, cDCs, and PDCs.
Figures
Figure 1.
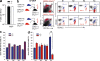
MDP and CDP share a similar phenotype. BM cells from _Cx3cr1_gfp/+ reporter mice were labeled with antibodies against lineage markers (CD11c, CD11b, NK1.1, CD3, Ter119, CD19, and Gr1), ckit (CD117), FLT3 (CD135), and CSF-1-R (CD115) and analyzed by FACS. CDP and MDP are overlapping populations; lineage− CD117low CD135+ CD115+ cells (CDP) express CX3CR1 (gfp), whereas CD117int CX3CR1 CD115+ cells (MDP) express CD135. Results are representative of >10 experiments.
Figure 2.
Differentiation potential of MDP in vivo. (a–c) MDPs from Cd45.1/2 or _Cd45.2 Cx3cr1_gfp/+ reporter mice were purified as described in Fig. S2 (available at
http://www.jem.org/cgi/content/full/jem.20081385/DC1
) and were adoptively transferred into irradiated (900 rad) C57BL/6 Cd45.2 congenic recipients. Spleens of recipient mice were analyzed at day 7 after transfer by flow cytometry, using lineage marker (NK1.1 CD3 CD19), CD11b, CD11c, and CD8-α antibodies. R1 corresponds to PDCs (Lin− CD11cint CD11b− CX3CR1+ cells), R2 and R3 correspond to cDCs (lin− CD11chigh CD11b− CX3CR1− cells and lin− CD11chigh CD11b+ CX3CR1+ cells), and R4 corresponds to monocytes (lin− CD11b+ CD11c− CX3CR1+ cells). The experiment was repeated five times with two to three mice per group and with similar results. (b) Donor-derived Lin− CD11cint CD11b− CX3CR1+ cells express PDCA1. (c) Role of anti-CD115 Ab. The number of donor-derived cells per spleen are represented after adoptive transfer of MDP, purified with or without CD115 antibody (n = at least 3 mice per group from two experiments). Error bars show SD. (d) The flow diagram represent the Lin− CD117int CD115+ CD135+ CX3CR1+ MDPs that give rise to monocytes, cDCs, and PDCs and their putative relationship with other myeloid precursors.
Figure 3.
CX3CR1 is important for the development of CD11b+ CD11c− monocytes. (a) MDP numbers in BM from CX3CR1+/− (black) and CX3CR1−/− (white) mice. Data are the mean ± SD of five mice per group. (b–d) Competitive adoptive transfer of MDP into irradiated host (900 rad). Equal numbers (104) of MDPs from Cd45.1/2 mice (blue) and Cd45.2/2 mice (red) were mixed and adoptively transferred into a Cd45.1 congenic recipient. Spleens of recipient mice were analyzed at day 7 after transfer by flow cytometry, using lineage marker (NK1.1 CD3 CD19), CD11b, CD11c, and CD8-α antibodies. (b and c) When both Cd45.1/2 and Cd45.2/2 donor mice were of the Cx3cr1+/− genotype, both donors contributed equally to NK1.1− CD3− CD19− CD11chigh CD11b− (R1, cDC) and NK1.1− CD3− CD19− CD11chigh CD11b+ (R2, cDC), NK1.1− CD3− CD19− CD11cint CD11b− (R3, PDC), and NK1.1− CD3− CD19− CD11b+ CD11c− splenocytes (R4). (b and d) In contrast, when Cd45.1/2 donor mice were of the Cx3cr1+/− genotype and Cd45.2/2 donor mice were of the Cx3cr1−/− genotype (CX3CR1 deficient), both donors contributed equally to CD11c+ cDCs in R1 and R2 but Cd45.1/2 Cx3cr1+/− MDPs were 10× more efficient than Cd45.2/2 Cx3cr1-deficient MDPs in generating CD11b+ CD11c− splenocytes in R3. Results in b are from one representative experiment out of three, with two to three mice per experimental group, and bar graphs in c and d represent the mean and SD from three independent experiments. The asterisk indicates a significative difference between groups (P < 0.05 using the Wilcoxon test).
Figure 4.
CX3CR1 is important for the recruitment of CD11b+ CD11c− monocytes to the spleen during infection. (a and b) Mouse fractalkine/CX3CL1 transcripts are detected in spleen tissue sections from WT mice around B cells follicles in the marginal zone/T cell area. Bars: (a) 500 µm; (b) 200 µm. (c) Short-term adoptive transfer of BM monocytes into irradiated WT recipient mice. Equal numbers (2 × 105) of BM monocytes from Cx3cr1+/− Cd45.1/2 mice (blue) and Cx3cr1−/− Cd45.2/2 mice (red) were mixed and adoptively transferred into a Cd45.1 congenic recipient. CX3CR1-expressing CD45.1/2 Gr1+ monocytes are recruited in the spleen of irradiated host 10× more efficiently than CX3CR1-deficient monocytes. The experiment was performed three times with two to three mice per group with similar results. (d–f) Monocyte recruitment in the spleen of infected mice. (d) BALB/c Cx3cr1+/− and Cx3cr1−/− mice (seven per group) were infected i.v. with live Lm (3 × 105), and monocytes (CX3CR1− gfp+ CD11b+ Ly-6C+ Ly-6G−, CD19−, CD3−, and NK1.1−) were enumerated after 24 h in the blood and spleen of infected and control uninfected mice. (e) C57BL/6 Cx3cr1+/− and Cx3cr1−/− mice (seven per group) were injected with 7 × 105 Lm, and monocytes were enumerated after 24 h in the blood and spleen of infected mice. (f) C57BL/6 Cx3cr1+/− and Cx3cr1−/− mice (seven per group) were injected with 7 × 103 Lm, and monocytes were enumerated after 48 h in the spleen of infected mice. Lm growth in the spleen of infected mice is shown. The asterisks indicate a significative difference between groups (P < 0.05). (g) Absence of proliferation of monocytes in the spleen of _Lm_-infected mice. BALB/c mice (n = 6 per group) were infected with 3 × 103 bacteria, spleen and leg bones were harvested 16 and 48 h later, and cells were processed as indicated in Fig. S5 (available at
http://www.jem.org/cgi/content/full/jem.20081385/DC1
). Data indicate the percentage of BM precursors and spleen monocytes in G0/G1 and in G2+S as analyzed by flow cytometry after DNA labeling with PI. The experiment was performed three times with similar results. (h) BALB/c Cx3cr1+/+ and Cx3cr1−/− mice were infected i.v. with live Lm (3 × 103). Data show the number of bacteria (mean ± SE) in the spleen 24 h after infection. (i) C57BL/6 Cx3cr1+/+ and Cx3cr1−/− mice were injected with 104 Lm. Data show the number of bacteria (mean ± SE) in the spleen 24 h after infection. Circles represent individual mice.
Figure 5.
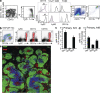
Functions of spleen CD115+ Gr1+/TipDCs. (a) CD11b+ spleen cells from Cx3cr1gfp/+ reporter mice negative for lineage marker (NK1.1, CD3, B220, CD19, and CD11c) were analyzed for Ly6G, Ly6C, F4/80, CD115, and Cx3CR1 (gfp) expression by multicolor flow cytometry. (b) TNF production by lineage− CD11clow CD11b+ cells 24 h after i.v. infection with Lm. Gated cells (circled) are displayed in red. Data show representative FACS profiles from three experiments. (c and d) Production of TNF-α, iNOS, and ROI by lineage− CD11clow CD11b+ Gr1+/Ly6C+ spleen cells from C57BL/6 mice 24 (c) and 48 h (d) after i.v. infection with 7 × 105 bacteria and 7 × 103 bacteria, respectively. The experiment was repeated five times with similar results. Error bars show SD. (e) iNOS+ cells in the spleen of BALB/c mice 24 h after infection with 105 WT Lm (red) express CD11b and Ly6C and are found in the perifollicular area and T cell area. B cell follicles are labeled with anti-B220 antibodies (green) and T cells are labeled with anti-CD3 antibodies (blue). Bar, 100 µm. The experiment was repeated five times with similar results.
Similar articles
- In vivo analysis of dendritic cell development and homeostasis.
Liu K, Victora GD, Schwickert TA, Guermonprez P, Meredith MM, Yao K, Chu FF, Randolph GJ, Rudensky AY, Nussenzweig M. Liu K, et al. Science. 2009 Apr 17;324(5925):392-7. doi: 10.1126/science.1170540. Epub 2009 Mar 12. Science. 2009. PMID: 19286519 Free PMC article. - Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor.
Sathe P, Metcalf D, Vremec D, Naik SH, Langdon WY, Huntington ND, Wu L, Shortman K. Sathe P, et al. Immunity. 2014 Jul 17;41(1):104-15. doi: 10.1016/j.immuni.2014.05.020. Immunity. 2014. PMID: 25035955 - Origin of the lamina propria dendritic cell network.
Bogunovic M, Ginhoux F, Helft J, Shang L, Hashimoto D, Greter M, Liu K, Jakubzick C, Ingersoll MA, Leboeuf M, Stanley ER, Nussenzweig M, Lira SA, Randolph GJ, Merad M. Bogunovic M, et al. Immunity. 2009 Sep 18;31(3):513-25. doi: 10.1016/j.immuni.2009.08.010. Epub 2009 Sep 10. Immunity. 2009. PMID: 19733489 Free PMC article. - The steady-state development of splenic dendritic cells.
Sathe P, Shortman K. Sathe P, et al. Mucosal Immunol. 2008 Nov;1(6):425-31. doi: 10.1038/mi.2008.56. Epub 2008 Sep 10. Mucosal Immunol. 2008. PMID: 19079209 Review. - Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses.
Geissmann F, Auffray C, Palframan R, Wirrig C, Ciocca A, Campisi L, Narni-Mancinelli E, Lauvau G. Geissmann F, et al. Immunol Cell Biol. 2008 Jul;86(5):398-408. doi: 10.1038/icb.2008.19. Epub 2008 Apr 8. Immunol Cell Biol. 2008. PMID: 18392044 Review.
Cited by
- Functions of Murine Dendritic Cells.
Durai V, Murphy KM. Durai V, et al. Immunity. 2016 Oct 18;45(4):719-736. doi: 10.1016/j.immuni.2016.10.010. Immunity. 2016. PMID: 27760337 Free PMC article. Review. - Inflammatory monocytes and the pathogenesis of viral encephalitis.
Terry RL, Getts DR, Deffrasnes C, van Vreden C, Campbell IL, King NJ. Terry RL, et al. J Neuroinflammation. 2012 Dec 17;9:270. doi: 10.1186/1742-2094-9-270. J Neuroinflammation. 2012. PMID: 23244217 Free PMC article. Review. - Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal.
Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, Hedrick CC, Cook HT, Diebold S, Geissmann F. Carlin LM, et al. Cell. 2013 Apr 11;153(2):362-75. doi: 10.1016/j.cell.2013.03.010. Cell. 2013. PMID: 23582326 Free PMC article. - Regulation of macrophage and dendritic cell responses by their lineage precursors.
Cortez-Retamozo V, Etzrodt M, Pittet MJ. Cortez-Retamozo V, et al. J Innate Immun. 2012;4(5-6):411-23. doi: 10.1159/000335733. Epub 2012 Mar 16. J Innate Immun. 2012. PMID: 22433183 Free PMC article. Review. - Chemokines and NK cells: regulators of development, trafficking and functions.
Bernardini G, Gismondi A, Santoni A. Bernardini G, et al. Immunol Lett. 2012 Jul 30;145(1-2):39-46. doi: 10.1016/j.imlet.2012.04.014. Immunol Lett. 2012. PMID: 22698182 Free PMC article. Review.
References
- Sasmono R.T., Oceandy D., Pollard J.W., Tong W., Pavli P., Wainwright B.J., Ostrowski M.C., Himes S.R., Hume D.A. 2003. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse.Blood. 101:1155–1163 - PubMed
- MacDonald K.P., Rowe V., Bofinger H.M., Thomas R., Sasmono T., Hume D.A., Hill G.R. 2005. The colony-stimulating factor 1 receptor is expressed on dendritic cells during differentiation and regulates their expansion.J. Immunol. 175:1399–1405 - PubMed
- Banchereau J., Steinman R.M. 1998. Dendritic cells and the control of immunity.Nature. 392:245–252 - PubMed
- Gordon S. 2002. Pattern recognition receptors: doubling up for the innate immune response.Cell. 111:927–930 - PubMed
- Taylor P.R., Gordon S. 2003. Monocyte heterogeneity and innate immunity.Immunity. 19:2–4 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous