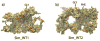Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics - PubMed (original) (raw)
Transport-related structures and processes of the nuclear pore complex studied through molecular dynamics
Lingling Miao et al. Structure. 2009.
Abstract
Nuclear pore complexes (NPCs) are selectively gated pathways between nucleoplasm and cytoplasm. Whereas small molecules can diffuse freely through NPCs, large molecules (>40 kD) can pass only when bound to transport receptors. The NPC central channel is filled with disordered proteins, rich in phenylalanine-glycine (FG) repeats, referred to as FG-nups. Our simulations, carried out at coarse-grained and all-atom levels, show that arrays of FG-nups tethered to a planar surface, at an FG-repeat density found in the NPC, form dynamic brush-like structures of multiprotein bundles, whereas individual FG-nups form dynamic globular structures. More than half of the FG-repeats are found on the surface of the bundles, offering a favorable environment for transport receptors. Binding to FG-repeats and a sliding motion of NTF2 induced by binding and unbinding to phenylalanines were observed when adding this transport receptor into one of the brush-like structures.
Figures
Figure 1
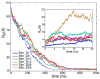
Coiling of individual, initially fully extended, nsp1 segments. The time evolution of the radius of gyration (Rg) is shown for 1 _μ_s CG simulations. The inset shows the time evolution of Rg during subsequent AA simulations, that started from the reverse-coarse-grained AA structures of the final (1 _μ_s) CG structures.
Figure 2
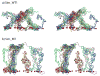
Initial configuration of the wildtype nsp1 array system (WT_array). a) Top view of the array. Only the N-terminus constrained to the substrate is shown for each chain. b) Side view. Each segment is shown in a different color with its constrained N-terminus represented by a red sphere. The water box including the array is shown in light purple and its dimension is labeled. The central segment of the array is indicated by a black circle.
Figure 3
Formation of brush-like structures. a) Time evolution of the brush-height for Sim_WT1 (red line), Sim_WT2 (blue line), and Sim_MT (green line) during 4 _μ_s CG simulations, with the inset showing the time evolution of the brush-height during the AA simulations. b) Pair distribution function g(r) of the FG-repeats of simulations Sim_WT1 (red line), Sim_WT2 (blue line), and Sim_MT (green line) averaged over the last 2 ns of their 10 ns AA simulations. The inset shows a histogram of the distances between adjacent binding spots on transport receptors (Isgro and Schulten, 2007b).
Figure 4
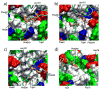
Stereo images of the brush-like structures of Sim_WT1 (a) and of Sim_MT (b). The CG brush-like structures of Sim_WT1 and Sim_MT at the end of their 4 _μ_s CG simulations are shown with each segment depicted in a different color; the constrained N-termini (tethering points) are represented by red spheres. Note that only a single periodic cell is shown; bundles at the cell boundary are actually engaged in contacts with segments of neighboring periodic cells, that are not shown. A large array of segments that includes three periodic cells is shown in Supplementary Materials Fig. S1.
Figure 5
Final brush-like structures for Sim_WT1 (a) and Sim_WT2 (b). The structures of Sim_WT1 and Sim_WT2 at the end of their AA simulations are shown in surface representation with the FG-repeats colored in orange and green. Many FG repeats are exposed to water and ready to bind to transport receptors. The final structures are dynamic.
Figure 6
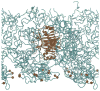
Side view of system B_NTF2 with NTF2 embedded. Only part of the system is shown with nsp1 segments in cyan, their constrained N-terminus C_α_ atoms as red spheres, and the embedded NTF2 in brown. Water and ions are not shown.
Figure 7
FG-repeat binding to NTF2. a) Binding of PHE343 to NTF2 binding spot 2 at 20 ns of Sim_NTF2. b) Binding of PHE343 and PHE341 to NTF2 binding spot 2 at 30 ns of Sim_NTF2. c) Binding of PHE341 to NTF2 binding spot 2 at the end of Sim_NTF2. d) Binding of PHE360 to NTF2 binding spot 1.3 at the end of Sim_NTF2. The NTF2 surface is colored according to residue type with polar basic residues colored in blue, polar acidic residues colored in red, polar neutral residues colored in green, and non-polar residues colored in white. Binding spots 2 and 1.3 are defined in (Isgro and Schulten, 2007a).
Figure 8
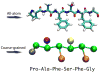
Nsp1 sequence (PAFSFG) in all-atom (AA, top) and in the equivalent coarse-grained (CG, bottom) description. The CG representation is shown with backbone beads in green, proline, alanine, phenylalanine and serine sidechains in tan, purple, orange and yellow, respectively. Glycine is represented by only a backbone bead.
Similar articles
- Assembly of Nsp1 nucleoporins provides insight into nuclear pore complex gating.
Gamini R, Han W, Stone JE, Schulten K. Gamini R, et al. PLoS Comput Biol. 2014 Mar 13;10(3):e1003488. doi: 10.1371/journal.pcbi.1003488. eCollection 2014 Mar. PLoS Comput Biol. 2014. PMID: 24626154 Free PMC article. - Crowding-induced phase separation of nuclear transport receptors in FG nucleoporin assemblies.
Davis LK, Ford IJ, Hoogenboom BW. Davis LK, et al. Elife. 2022 Jan 31;11:e72627. doi: 10.7554/eLife.72627. Elife. 2022. PMID: 35098921 Free PMC article. - The Effect of FG-Nup Phosphorylation on NPC Selectivity: A One-Bead-Per-Amino-Acid Molecular Dynamics Study.
Mishra A, Sipma W, Veenhoff LM, Van der Giessen E, Onck PR. Mishra A, et al. Int J Mol Sci. 2019 Jan 30;20(3):596. doi: 10.3390/ijms20030596. Int J Mol Sci. 2019. PMID: 30704069 Free PMC article. - Deciphering the intrinsically disordered characteristics of the FG-Nups through the lens of polymer physics.
Matsuda A, Mansour A, Mofrad MRK. Matsuda A, et al. Nucleus. 2024 Dec;15(1):2399247. doi: 10.1080/19491034.2024.2399247. Epub 2024 Sep 16. Nucleus. 2024. PMID: 39282864 Free PMC article. Review. - Biomechanics of the transport barrier in the nuclear pore complex.
Stanley GJ, Fassati A, Hoogenboom BW. Stanley GJ, et al. Semin Cell Dev Biol. 2017 Aug;68:42-51. doi: 10.1016/j.semcdb.2017.05.007. Epub 2017 May 12. Semin Cell Dev Biol. 2017. PMID: 28506890 Review.
Cited by
- The yeast nuclear pore complex and transport through it.
Aitchison JD, Rout MP. Aitchison JD, et al. Genetics. 2012 Mar;190(3):855-83. doi: 10.1534/genetics.111.127803. Genetics. 2012. PMID: 22419078 Free PMC article. - Brownian dynamics simulation of nucleocytoplasmic transport: a coarse-grained model for the functional state of the nuclear pore complex.
Moussavi-Baygi R, Jamali Y, Karimi R, Mofrad MR. Moussavi-Baygi R, et al. PLoS Comput Biol. 2011 Jun;7(6):e1002049. doi: 10.1371/journal.pcbi.1002049. Epub 2011 Jun 2. PLoS Comput Biol. 2011. PMID: 21673865 Free PMC article. - How sequence determines elasticity of disordered proteins.
Cheng S, Cetinkaya M, Gräter F. Cheng S, et al. Biophys J. 2010 Dec 15;99(12):3863-9. doi: 10.1016/j.bpj.2010.10.011. Biophys J. 2010. PMID: 21156127 Free PMC article. - Calcium regulation of nucleocytoplasmic transport.
Sarma A, Yang W. Sarma A, et al. Protein Cell. 2011 Apr;2(4):291-302. doi: 10.1007/s13238-011-1038-x. Epub 2011 Apr 27. Protein Cell. 2011. PMID: 21528351 Free PMC article. Review. - Deciphering the Structure and Function of Nuclear Pores Using Single-Molecule Fluorescence Approaches.
Musser SM, Grünwald D. Musser SM, et al. J Mol Biol. 2016 May 22;428(10 Pt A):2091-119. doi: 10.1016/j.jmb.2016.02.023. Epub 2016 Mar 2. J Mol Biol. 2016. PMID: 26944195 Free PMC article. Review.
References
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
- Alexander S. Polymer adsorption on small spheres. a scaling approach. J Phys France. 1977;38:977–981.
- Bayliss R, Littlewood T, Strawn LA, Wente SR, Stewart M. GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J Biol Chem. 2002b;277:50597–50606. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067887-05/GM/NIGMS NIH HHS/United States
- R01-GM067887/GM/NIGMS NIH HHS/United States
- P41 RR005969/RR/NCRR NIH HHS/United States
- R01 GM067887/GM/NIGMS NIH HHS/United States
- P41-RR05969/RR/NCRR NIH HHS/United States
- P41 RR005969-19/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources