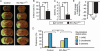Rac1 is a critical mediator of endothelium-derived neurotrophic activity - PubMed (original) (raw)
Comparative Study
Rac1 is a critical mediator of endothelium-derived neurotrophic activity
Naoki Sawada et al. Sci Signal. 2009.
Abstract
The therapeutic potential of neurotrophic factors has been hampered by their inability to achieve adequate tissue penetration. Brain blood vessels, however, could be an alternative target for neurosalvage therapies by virtue of their close proximity to neurons. Here we show that hemizygous deletion of Rac1 in mouse endothelial cells (ECs) attenuates brain injury and edema after focal cerebral ischemia. Microarray analysis of Rac1(+/-) ECs revealed enrichment of stress response genes, basement membrane components, and neurotrophic factors that could affect neuronal survival. Consistent with these expression profiles, endothelial Rac1 hemizygosity enhanced antioxidative and endothelial barrier capacities and potentiated paracrine neuroprotective activities through the up-regulation of the neurotrophic factor, artemin. Endothelial Rac1, therefore, could be an important therapeutic target for promoting endothelial barrier integrity and neurotrophic activity.
Figures
Fig. 1
Haploinsufficiency of endothelial Rac1 in mice leads to neuroprotection. EC-Rac1+/− and control mice underwent 2-hour MCAo followed by 22-hour reperfusion. (A) Representative brain sections stained with 2,3,5-triphenyltetrazolium chloride visualizing the infarct area. (B and C) Infarct (B) and edema (C) volumes of the ischemic hemisphere. (D) Neurological deficit score (n = 19 to 23 mice; *P < 0.05, Mann-Whitney test). (E) Change in CBF in the ischemic hemisphere at the end of 2-hour MCAo. (B, C, and E) Values are percent of the nonischemic hemisphere and shown as means ± SEM of 7 to 10 mice (*P < 0.05, ANOVA). NS, not significant.
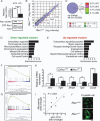
Fig. 2
Expression profiling of Rac1+/− ECs. Differential mRNA expression between Rac1+/− and Rac1+/+ mouse ECs was explored by microarray analysis. (A) qRT-PCR, immunoblotting, and GST-PAK (glutathione _S_-transferase–p21-activated protein kinase) pull-down assays showing Rac1 expression and activity in Rac1+/− and Rac1+/+ ECs. (B and C) The up-regulated (red) and down-regulated (green) genes in Rac1+/− ECs. The scatter plot (B) shows 12,089 gene features with the signal intensity >200. (D and E) Functional annotation clustering of the down-regulated (D) and up-regulated (E) genes. (F and G) GSEA showing enrichment of cell cycle–related genes in Rac1+/+ (F) and TGF-β–inducible genes in Rac1+/− ECs (G). Each plot displays (top) progression of the running enrichment score; (middle) “hits” in the gene set against the ranked list of all genes in the data; (bottom) histogram for the ranked list. (H and I) qRT-PCR (H) and correlation analysis (I) to validate the microarray data. (J) Crystallin αB immunostaining of Rac1+/− versus Rac1+/+ ECs. Scale bar, 25 μm. Values are means ± SEM of three to four replicates (*P < 0.05, **P < 0.01; ANOVA).
Fig. 3
Phenotypic profiling of Rac1+/− ECs. (A, B, D, E, and F) Rac1+/+ and Rac1+/− mouse ECs were sequentially exposed to 4-hour hypoxia and 2-hour (A and D) or 20-hour (B, E, and F) reoxygenation (H/R) or treated with normoxia (Norm). (A) NADPH oxidase activity (left) and ROS production (right) as determined by DPI-inhibitable lucigenin chemiluminescence and DCF fluorescence, respectively. (B) Apoptotic death was assessed by flow cytometry (left) as the Annexin V–EGFP–labeled EC fraction (right). (C)ECs at subconfluence (Sub) or prolonged confluence (>4 weeks) (Sheet) were subjected to standard trypsinization until single-cell suspensions were obtained. (D) The permeability of confluent EC monolayers grown on 0.4-μm Transwell inserts was assessed as HRP leak across the membrane during normoxia and H/R. (E) After exposure to normoxia or H/R, ECs were co-cultured with [3H]thymidine-labeled monocytes (THP-1) for 3 hours. After washing, monocyte-EC binding was assessed as the cell lysate radioactivity. (F) VCAM-1 mRNA abundance was determined by qRT-PCR. (A, C to F) Values are means ± SEM of triplicates (*P < 0.05, **P < 0.01; ANOVA).
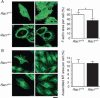
Fig. 4
Decreased actin polymerization and circumferential redistribution of F-actin to the cortical areas in Rac1+/− ECs. Mouse ECs cultured in the presence of 20% serum were fixed and stained with (A) phalloidin to visualize F-actin and (B) antibody against β-tubulin for microtubules (MT). The F-actin and MT-labeled areas were quantified and represented as percent of total cell areas. Scale bar, 25 μm. Values are means ± SEM of five to seven replicates (*P < 0.05; ANOVA).
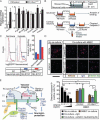
Fig. 5
Rac1+/− ECs show enhanced neurotrophic activity through paracrine mechanisms. (A) mRNA expression of the potential neurotrophic factors was assessed in the EC-Rac1+/− and control mouse whole brains by qRT-PCR (n = 7 mouse brains). (B) Schematic of EC-neuron co-culture. (C) Co-culture with Rac1+/− versus Rac1+/+ ECs mitigates hypoxia-induced apoptotic death of SH-SY5Y cells, as assessed by Annexin V-EGFP labeling and flow cytometry. (D) Cortical neurons isolated from fetal mouse brains (∼E16) were co-cultured with EC-Rac1+/− or control MBECs in the presence of artemin-neutralizing antibody (Ab), control IgG, or vehicle in the lower chamber. Control neurons were treated with blank inserts and recombinant artemin proteins. Upper panel, after OGD and reoxygenation, the neurons were double-labeled with DAPI (blue) and cleaved caspase 3 (red). Scale bar, 1 mm. Lower panel, quantification of the cleaved caspase 3–positive fractions. (E) A proposed model of neuroprotection through targeting endothelial Rac1. Values are means ± SEM (*P < 0.05, **P < 0.01; ANOVA).
Similar articles
- Genetic deletion of Rac1 GTPase reveals its critical role in actin stress fiber formation and focal adhesion complex assembly.
Guo F, Debidda M, Yang L, Williams DA, Zheng Y. Guo F, et al. J Biol Chem. 2006 Jul 7;281(27):18652-9. doi: 10.1074/jbc.M603508200. Epub 2006 May 11. J Biol Chem. 2006. PMID: 16698790 - The role of VASP in regulation of cAMP- and Rac 1-mediated endothelial barrier stabilization.
Schlegel N, Burger S, Golenhofen N, Walter U, Drenckhahn D, Waschke J. Schlegel N, et al. Am J Physiol Cell Physiol. 2008 Jan;294(1):C178-88. doi: 10.1152/ajpcell.00273.2007. Epub 2007 Nov 7. Am J Physiol Cell Physiol. 2008. PMID: 17989211 - Neuronal Rho GTPase Rac1 elimination confers neuroprotection in a mouse model of permanent ischemic stroke.
Karabiyik C, Fernandes R, Figueiredo FR, Socodato R, Brakebusch C, Lambertsen KL, Relvas JB, Santos SD. Karabiyik C, et al. Brain Pathol. 2018 Jul;28(4):569-580. doi: 10.1111/bpa.12562. Epub 2017 Oct 24. Brain Pathol. 2018. PMID: 28960571 Free PMC article. - Epidermal deletion of Rac1 causes stem cell depletion, irrespective of whether deletion occurs during embryogenesis or adulthood.
Benitah SA, Watt FM. Benitah SA, et al. J Invest Dermatol. 2007 Jun;127(6):1555-7. doi: 10.1038/sj.jid.5700738. Epub 2007 Feb 15. J Invest Dermatol. 2007. PMID: 17301832 No abstract available. - CD44 regulates hepatocyte growth factor-mediated vascular integrity. Role of c-Met, Tiam1/Rac1, dynamin 2, and cortactin.
Singleton PA, Salgia R, Moreno-Vinasco L, Moitra J, Sammani S, Mirzapoiazova T, Garcia JG. Singleton PA, et al. J Biol Chem. 2007 Oct 19;282(42):30643-57. doi: 10.1074/jbc.M702573200. Epub 2007 Aug 16. J Biol Chem. 2007. PMID: 17702746
Cited by
- High-Frequency Irreversible Electroporation (H-FIRE) Induced Blood-Brain Barrier Disruption Is Mediated by Cytoskeletal Remodeling and Changes in Tight Junction Protein Regulation.
Partridge BR, Kani Y, Lorenzo MF, Campelo SN, Allen IC, Hinckley J, Hsu FC, Verbridge SS, Robertson JL, Davalos RV, Rossmeisl JH. Partridge BR, et al. Biomedicines. 2022 Jun 11;10(6):1384. doi: 10.3390/biomedicines10061384. Biomedicines. 2022. PMID: 35740406 Free PMC article. - Endothelial PGC-1α mediates vascular dysfunction in diabetes.
Sawada N, Jiang A, Takizawa F, Safdar A, Manika A, Tesmenitsky Y, Kang KT, Bischoff J, Kalwa H, Sartoretto JL, Kamei Y, Benjamin LE, Watada H, Ogawa Y, Higashikuni Y, Kessinger CW, Jaffer FA, Michel T, Sata M, Croce K, Tanaka R, Arany Z. Sawada N, et al. Cell Metab. 2014 Feb 4;19(2):246-58. doi: 10.1016/j.cmet.2013.12.014. Cell Metab. 2014. PMID: 24506866 Free PMC article. - Novel aspects of the roles of Rac1 GTPase in the cardiovascular system.
Sawada N, Li Y, Liao JK. Sawada N, et al. Curr Opin Pharmacol. 2010 Apr;10(2):116-21. doi: 10.1016/j.coph.2009.11.004. Epub 2010 Jan 7. Curr Opin Pharmacol. 2010. PMID: 20060361 Free PMC article. Review. - Activation of endothelial ras-related C3 botulinum toxin substrate 1 (Rac1) improves post-stroke recovery and angiogenesis via activating Pak1 in mice.
Bu F, Min JW, Munshi Y, Lai YJ, Qi L, Urayama A, McCullough LD, Li J. Bu F, et al. Exp Neurol. 2019 Dec;322:113059. doi: 10.1016/j.expneurol.2019.113059. Epub 2019 Sep 6. Exp Neurol. 2019. PMID: 31499064 Free PMC article. Review. - Role of reactive oxygen and nitrogen species in the vascular responses to inflammation.
Kvietys PR, Granger DN. Kvietys PR, et al. Free Radic Biol Med. 2012 Feb 1;52(3):556-592. doi: 10.1016/j.freeradbiomed.2011.11.002. Epub 2011 Nov 12. Free Radic Biol Med. 2012. PMID: 22154653 Free PMC article. Review.
References
- Cheng T, Liu D, Griffin JH, Fernandez JA, Castellino F, Rosen ED, Fukudome K, Zlokovic BV. Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat. Med. 2003;9:338–342. - PubMed
- Abbott NJ, Ronnback L, Hansson E. Astrocyte–endothelial interactions at the blood–brain barrier. Nat. Rev. Neurosci. 2006;7:41–53. - PubMed
- Cunningham LA, Wetzel M, Rosenberg GA. Multiple roles for MMPs and TIMPs in cerebral ischemia. Glia. 2005;50:329–339. - PubMed
- Gregg D, Rauscher FM, Goldschmidt-Clermont PJ. Rac regulates cardiovascular superoxide through diverse molecular interactions: More than a binary GTP switch. Am. J. Physiol. Cell Physiol. 2003;285:C723–C734. - PubMed
- Tzima E. Role of small GTPases in endothelial cytoskeletal dynamics and the shear stress response. Circ. Res. 2006;98:176–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL052233-12/HL/NHLBI NIH HHS/United States
- R01 HL070274-05/HL/NHLBI NIH HHS/United States
- R01 HL052233-05/HL/NHLBI NIH HHS/United States
- R01 HL052233-09/HL/NHLBI NIH HHS/United States
- R01 DK062729-05/DK/NIDDK NIH HHS/United States
- R01 HL052233-10/HL/NHLBI NIH HHS/United States
- R01 HL080187/HL/NHLBI NIH HHS/United States
- R01 HL052233-11/HL/NHLBI NIH HHS/United States
- R01 HL080187-04/HL/NHLBI NIH HHS/United States
- R01 HL070274-04/HL/NHLBI NIH HHS/United States
- R01 HL080187-02/HL/NHLBI NIH HHS/United States
- R01 HL052233/HL/NHLBI NIH HHS/United States
- R01 DK062729/DK/NIDDK NIH HHS/United States
- R01 HL080187-03/HL/NHLBI NIH HHS/United States
- R01 HL070274/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials