An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network - PubMed (original) (raw)
An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network
Lena Ho et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Distinctive SWI/SNF-like ATP-dependent chromatin remodeling esBAF complexes are indispensable for the maintenance and pluripotency of mouse embryonic stem (ES) cells [Ho L, et al. (2009) Proc Natl Acad Sci USA 10.1073/pnas.0812889106]. To understand the mechanism underlying the roles of these complexes in ES cells, we performed high-resolution genome-wide mapping of the core ATPase subunit, Brg, using ChIP-Seq technology. We find that esBAF, as represented by Brg, binds to genes encoding components of the core ES transcriptional circuitry, including Polycomb group proteins. esBAF colocalizes extensively with transcription factors Oct4, Sox2 and Nanog genome-wide, and shows distinct functional interactions with Oct4 and Sox2 at its target genes. Surprisingly, no significant colocalization of esBAF with PRC2 complexes, represented by Suz12, is observed. Lastly, esBAF colocalizes with Stat3 and Smad1 genome-wide, consistent with a direct and critical role in LIF and BMP signaling for maintaining self-renewal. Taken together, our studies indicate that esBAF is an essential component of the core pluripotency transcriptional network, and might also be a critical component of the LIF and BMP signaling pathways essential for maintenance of self-renewal and pluripotency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
High-resolution genome-wide analysis of esBAF occupancy. (A) (Left) Specificity of J1 antibody used for ChIP-Seq both by immunoblotting of ES nuclear extracts and immunoprecipitation of BAF complexes from ES extracts and visualization by silver stain. (Right) Distribution of Brg-bound regions throughout the genome. TSS, transcription start site; genic regions are defined as 5 Kb downstream of TSS to the end of an annotated gene. (TES; transcription end site.) (B) Average distribution of Brg-normalized tag density across a gene unit (Left). Higher-resolution analysis of average tag density surrounding the TSS (Right). In each plot, genes were classified into 10 groups based on expression levels in ES cells (highest to lowest represented by colored lines), and average tag density across the gene unit was plotted for each group.
Fig. 2.
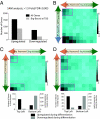
esBAF complexes occupy genes of the core transcription network. (A) All Brg-bound genes were placed in a matrix to determine their expression in ES cells (x axis), and whether they are ES-specific or differentiation-specific according to their direction of regulation as ES cells differentiate to day 14 embryoid bodies (embryoid/ES fold change). Intensity of the heat map represents the absolute numbers of genes falling in each square of the matrix, as indicated by color bar on the right. In this matrix, y-axis represents n = 17,030 genes that had unique transcripts represented by probes on Affymetrix's MOE430_2 expression arrays minus ∼15% genes with low expression levels in both ES and differentiated cells, and x-axis represents 5,630 genes that are bound by Brg (see SI Methods). (B) Brg occupancy on genes of the core ES cell circuitry from the University of California, Santa Cruz (UCSC) genome browser. Brg ChIP (Left) and IgG ChIP (Right) plotted on comparable tag density axes.
Fig. 3.
Brg represses developmental genes and refines ES-specific genes by cooperating with Oct4 and Sox2. (A) SAM analysis of microarray data from Brg knockdown (KD) ES cells compared with control knockdown, 96 h after transfection of constructs. Genes that were significantly changed were further classified as Brg-bound within the gene unit (gray bars) or not (black bars). (B) Brg-bound genes were placed in a matrix with fold change after Brg KD on the x axis against EB/ES fold (i.e., differentiation- or ES-specific) on the y axis to ascertain whether Brg-dependent target genes are ES-specific, or differentiation associated, and in which direction they are changed upon Brg KD. (C) Brg-bound genes were placed in a matrix with fold change after Brg KD on the x axis against observed fold change of the same genes afternOct4 KD (25) or Sox2 KO (D) (12) in ES cells on the y axis to determine the functional interaction between Oct4/Sox2 and Brg. Bar graphs indicate the natural developmental fate of genes within the top left (solid) squares and bottom left (dashed) squares by comparing their fold change between ES cells and day 14 embryoid bodies.
Fig. 4.
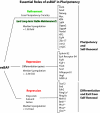
Proposed roles of esBAFs in the regulation of the core circuitry. In one strategy esBAF functionally interacts with Sox2 and Oct4 to refine the levels of ES-specific genes, or to repress the expression of some differentiation genes. In a second strategy that probably facilitates exit from the pluripotent state, esBAF directly regulates the expression of some PcG proteins. The median fold change of genes in each group is noted. Each of the genes on the right are direct targets of esBAF complexes by ChIP-Seq. *, PcG genes that were ditectably upregulated by microarray analysis.
Similar articles
- An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency.
Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. Ho L, et al. Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):5181-6. doi: 10.1073/pnas.0812889106. Epub 2009 Mar 11. Proc Natl Acad Sci U S A. 2009. PMID: 19279220 Free PMC article. - Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells.
Kidder BL, Yang J, Palmer S. Kidder BL, et al. PLoS One. 2008;3(12):e3932. doi: 10.1371/journal.pone.0003932. Epub 2008 Dec 11. PLoS One. 2008. PMID: 19079543 Free PMC article. - SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells.
Kidder BL, Palmer S, Knott JG. Kidder BL, et al. Stem Cells. 2009 Feb;27(2):317-28. doi: 10.1634/stemcells.2008-0710. Stem Cells. 2009. PMID: 19056910 - Transcriptional regulatory networks in embryonic stem cells.
Chen X, Vega VB, Ng HH. Chen X, et al. Cold Spring Harb Symp Quant Biol. 2008;73:203-9. doi: 10.1101/sqb.2008.73.026. Epub 2008 Nov 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19022762 Review. - The transcriptional network controlling pluripotency in ES cells.
Orkin SH, Wang J, Kim J, Chu J, Rao S, Theunissen TW, Shen X, Levasseur DN. Orkin SH, et al. Cold Spring Harb Symp Quant Biol. 2008;73:195-202. doi: 10.1101/sqb.2008.72.001. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19478325 Review.
Cited by
- Dynamic regulation of epigenomic landscapes during hematopoiesis.
Abraham BJ, Cui K, Tang Q, Zhao K. Abraham BJ, et al. BMC Genomics. 2013 Mar 19;14:193. doi: 10.1186/1471-2164-14-193. BMC Genomics. 2013. PMID: 23510235 Free PMC article. - ARID1A-BAF coordinates ZIC2 genomic occupancy for epithelial-to-mesenchymal transition in cranial neural crest specification.
Barnada SM, Giner de Gracia A, Morenilla-Palao C, López-Cascales MT, Scopa C, Waltrich FJ Jr, Mikkers HMM, Cicardi ME, Karlin J, Trotti D, Peterson KA, Brugmann SA, Santen GWE, McMahon SB, Herrera E, Trizzino M. Barnada SM, et al. Am J Hum Genet. 2024 Oct 3;111(10):2232-2252. doi: 10.1016/j.ajhg.2024.07.022. Epub 2024 Sep 2. Am J Hum Genet. 2024. PMID: 39226899 Free PMC article. - Genome-wide nucleosome positioning during embryonic stem cell development.
Teif VB, Vainshtein Y, Caudron-Herger M, Mallm JP, Marth C, Höfer T, Rippe K. Teif VB, et al. Nat Struct Mol Biol. 2012 Nov;19(11):1185-92. doi: 10.1038/nsmb.2419. Epub 2012 Oct 21. Nat Struct Mol Biol. 2012. PMID: 23085715 - Stemness factor Sall4 is required for DNA damage response in embryonic stem cells.
Xiong J, Todorova D, Su NY, Kim J, Lee PJ, Shen Z, Briggs SP, Xu Y. Xiong J, et al. J Cell Biol. 2015 Mar 2;208(5):513-20. doi: 10.1083/jcb.201408106. J Cell Biol. 2015. PMID: 25733712 Free PMC article. - Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation.
Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. Melcer S, et al. Nat Commun. 2012 Jun 19;3:910. doi: 10.1038/ncomms1915. Nat Commun. 2012. PMID: 22713752 Free PMC article.
References
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. - PubMed
- Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. - PubMed
- Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NS046789/NS/NINDS NIH HHS/United States
- R01 NS046789/NS/NINDS NIH HHS/United States
- R01 HD055391/HD/NICHD NIH HHS/United States
- R37 NS046789/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- ImNIH/Intramural NIH HHS/United States
- HD55391/HD/NICHD NIH HHS/United States
- AI060037/AI/NIAID NIH HHS/United States
- R01 AI060037/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous