Structural analysis of an open active site conformation of nonheme iron halogenase CytC3 - PubMed (original) (raw)
Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
Cintyu Wong et al. J Am Chem Soc. 2009.
Abstract
CytC3, a member of the recently discovered class of nonheme Fe(II) and alpha-ketoglutarate (alphaKG)-dependent halogenases, catalyzes the double chlorination of L-2-aminobutyric acid (Aba) to produce a known Streptomyces antibiotic, gamma,gamma-dichloroaminobutyrate. Unlike the majority of the Fe(II)-alphaKG-dependent enzymes that catalyze hydroxylation reactions, halogenases catalyze a transfer of halides. To examine the important enzymatic features that discriminate between chlorination and hydroxylation, the crystal structures of CytC3 both with and without alphaKG/Fe(II) have been solved to 2.2 A resolution. These structures capture CytC3 in an open active site conformation, in which no chloride is bound to iron. Comparison of the open conformation of CytC3 with the closed conformation of another nonheme iron halogenase, SyrB2, suggests two important criteria for creating an enzyme-bound Fe-Cl catalyst: (1) the presence of a hydrogen-bonding network between the chloride and surrounding residues, and (2) the presence of a hydrophobic pocket in which the chloride resides.
Figures
Figure 1
Production of 4-Cl-
l
-Aba by CytC1−3. CytC1 loads
l
-Aba-AMP onto the phosphopantetheine arm (wavy line) on CytC2. CytC3 chlorinates the tethered substrate to form 4-Cl-
l
-Aba. CytC1 is an adenylation domain (A) and CytC2 is a thiolation domain (T).
Figure 2
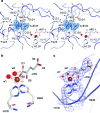
Overall structure of CytC3 dimer. (a) Crystallographic dimer is colored by molecule. The iron ligands: two protein residues (His118 and His240) and αKG are shown in stick representation, and waters in the active site are shown in a spherical representation. The ends of the disordered region are indicated as “missing loop”, which consists of residues 178−219. (b) Structural alignment of SyrB2 monomer (pink) and CytC3 dimer (blue). (c) Surface representation of CytC3 dimer showing access to active site on one face of the crystallographic dimer. Active site waters are colored in red; each monomer of CytC3 is colored yellow and blue. The ends of the disordered region are colored in green.
Figure 4
CytC3 active site region. (a) Open active site in CytC3 structure allows for molecules from crystallization solution to bind, shown in stereo view. A 2_F_o − _F_c composite omit map contoured at 1σ is shown in blue mesh around excess ligands (αKG and sulfate). Iron is shown in brown, and waters are shown in red. (b) An iron anomalous difference map contoured at 12σ is shown in brown mesh around the iron. The αKG is labeled with the numbering nomenclature for each oxygen atoms. (c) A 2_F_o − _F_c composite omit map contoured at 2σ is shown in blue mesh around the active site ligands.
Figure 5
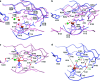
Active site comparison for CytC3 and SyrB2. Iron is shown in brown, chloride ion is shown in green, and waters are shown in red. (a) Comparison between CytC3′s “open” active site (blue) and SyrB2’s “closed” active site (pink), where structures are aligned by F106, K108, H125, W150, T148, R253, and R259 of CytC3 with the corresponding residues in SyrB2. (b) Comparison of CytC3 (blue) and SyrB2 (pink) where structures are aligned by the two active site histidines. (c) Active site residues of SyrB2 and relevant distances. Dash lines indicate hydrogen bonding with labeled distances. (d) Active site residues of CytC3 and relevant distances. The corresponding hydrogen bond interactions between the protein and αKG as observed in SyrB2 are indicated by black dashes. New interactions observed between CytC3 and αKG are indicated by green dashes with labeled distances.
Figure 6
Chloride binding pocket of SyrB2 and CytC3. Iron is shown in brown, chloride ion is shown in green, and waters are shown in red. (a) Residues A118, F121, and the β-carbon of S231 form a nice hydrophobic pocket for chloride in the closed state of SyrB2 active site. (b) The corresponding residues, A120, F123, and S236, in the open state of the CytC3 active site are too far from the chloride binding site to form a hydrophobic pocket environment.
Similar articles
- Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3.
Galonić DP, Barr EW, Walsh CT, Bollinger JM Jr, Krebs C. Galonić DP, et al. Nat Chem Biol. 2007 Feb;3(2):113-6. doi: 10.1038/nchembio856. Epub 2007 Jan 14. Nat Chem Biol. 2007. PMID: 17220900 - Structural and Functional Insights into a Nonheme Iron- and α-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Dai L, Zhang X, Hu Y, Shen J, Zhang Q, Zhang L, Min J, Chen CC, Liu Y, Huang JW, Guo RT. Dai L, et al. Appl Environ Microbiol. 2022 May 10;88(9):e0249721. doi: 10.1128/aem.02497-21. Epub 2022 Apr 18. Appl Environ Microbiol. 2022. PMID: 35435717 Free PMC article. - CD and MCD of CytC3 and taurine dioxygenase: role of the facial triad in alpha-KG-dependent oxygenases.
Neidig ML, Brown CD, Light KM, Fujimori DG, Nolan EM, Price JC, Barr EW, Bollinger JM Jr, Krebs C, Walsh CT, Solomon EI. Neidig ML, et al. J Am Chem Soc. 2007 Nov 21;129(46):14224-31. doi: 10.1021/ja074557r. Epub 2007 Oct 30. J Am Chem Soc. 2007. PMID: 17967013 Free PMC article. - Oxidative tailoring reactions catalyzed by nonheme iron-dependent enzymes: streptorubin B biosynthesis as an example.
Sydor PK, Challis GL. Sydor PK, et al. Methods Enzymol. 2012;516:195-218. doi: 10.1016/B978-0-12-394291-3.00002-2. Methods Enzymol. 2012. PMID: 23034230 Review. - Variations of the 2-His-1-carboxylate theme in mononuclear non-heme FeII oxygenases.
Straganz GD, Nidetzky B. Straganz GD, et al. Chembiochem. 2006 Oct;7(10):1536-48. doi: 10.1002/cbic.200600152. Chembiochem. 2006. PMID: 16858718 Review.
Cited by
- Selective Radical Transfer in a Series of Nonheme Iron(III) Complexes.
Yadav V, Wen L, Yadav S, Siegler MA, Goldberg DP. Yadav V, et al. Inorg Chem. 2023 Oct 30;62(43):17830-17842. doi: 10.1021/acs.inorgchem.3c02617. Epub 2023 Oct 19. Inorg Chem. 2023. PMID: 37857315 Free PMC article. - Naturally Occurring Organohalogen Compounds-A Comprehensive Review.
Gribble GW. Gribble GW. Prog Chem Org Nat Prod. 2023;121:1-546. doi: 10.1007/978-3-031-26629-4_1. Prog Chem Org Nat Prod. 2023. PMID: 37488466 Review. - Non-Native Anionic Ligand Binding and Reactivity in Engineered Variants of the Fe(II)- and α-Ketoglutarate-Dependent Oxygenase, SadA.
Chan NH, Gomez CA, Vennelakanti V, Du Q, Kulik HJ, Lewis JC. Chan NH, et al. Inorg Chem. 2022 Sep 12;61(36):14477-14485. doi: 10.1021/acs.inorgchem.2c02872. Epub 2022 Aug 31. Inorg Chem. 2022. PMID: 36044713 Free PMC article. - Determining the inherent selectivity for carbon radical hydroxylation versus halogenation with high-spin oxoiron(iv)-halide complexes: a concerted rebound step.
Tao Y, Li Z, Zhang Y, Sun K, Liu Z. Tao Y, et al. RSC Adv. 2022 Mar 29;12(16):9891-9897. doi: 10.1039/d2ra01384c. eCollection 2022 Mar 25. RSC Adv. 2022. PMID: 35424943 Free PMC article. - Halogen Transfer to Carbon Radicals by High-Valent Iron Chloride and Iron Fluoride Corroles.
Farley GW, Siegler MA, Goldberg DP. Farley GW, et al. Inorg Chem. 2021 Nov 15;60(22):17288-17302. doi: 10.1021/acs.inorgchem.1c02666. Epub 2021 Oct 28. Inorg Chem. 2021. PMID: 34709780 Free PMC article.
References
- Walsh C. Nature 2000, 406, 775–781. - PubMed
- Fenical W.; Jensen P. R. Nat. Chem. Biol. 2006, 2, 666–73. - PubMed
- Grgurina I.; Barca A.; Cervigni S.; Gallo M.; Scaloni A.; Pucci P. Experientia 1994, 50, 130–133. - PubMed
- Schnarr N. A.; Khosla C. Nature 2005, 436, 1094–1095. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM65337/GM/NIGMS NIH HHS/United States
- R01 GM049338/GM/NIGMS NIH HHS/United States
- GM49338/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 ES002109/ES/NIEHS NIH HHS/United States
- R01 GM065337/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical