Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis - PubMed (original) (raw)
Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis
Jessica H Fong et al. PLoS Comput Biol. 2009 Mar.
Abstract
We perform a large-scale study of intrinsically disordered regions in proteins and protein complexes using a non-redundant set of hundreds of different protein complexes. In accordance with the conventional view that folding and binding are coupled, in many of our cases the disorder-to-order transition occurs upon complex formation and can be localized to binding interfaces. Moreover, analysis of disorder in protein complexes depicts a significant fraction of intrinsically disordered regions, with up to one third of all residues being disordered. We find that the disorder in homodimers, especially in symmetrical homodimers, is significantly higher than in heterodimers and offer an explanation for this interesting phenomenon. We argue that the mechanisms of regulation of binding specificity through disordered regions in complexes can be as common as for unbound monomeric proteins. The fascinating diversity of roles of disordered regions in various biological processes and protein oligomeric forms shown in our study may be a subject of future endeavors in this area.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
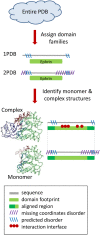
Figure 1. Flowchart showing the construction of the dataset.
Figure 2. Diagram illustrating the definition of conserved binding modes and construction of the test set.
Structures 1PDB , 2 PDB and 3PDB have a recurrent mode of interaction between families A and B, this constitutes CBM#1. Structures 4PDB and 5PDB use another binding mode which is also conserved between two different structures and therefore constitutes CBM#2. There is only one instance of binding mode between family A and C, therefore it is not a CBM. Structures of two different representatives of family A in complex and monomeric forms are shown. Interface regions mapped from complex to monomer are shown as orange arcs and disordered regions on the inferred interface regions are shown in orange, all other disordered regions are shown in yellow. Fraction disorder in family A in a complex state is calculated by averaging over all structures of a given CBM (1PDB, 2PDB, 3PDB for CBM#1 and 4PDB, 5PDB for CBM#2).
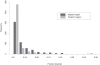
Figure 3. Histogram of fraction disorder in protein complexes (“test588”).
Fraction disorder is calculated for aligned (black) and footprint regions (grey). The “footprint region” extends from the first to the last residue in the alignment mapping CDD family to a given chain. “Aligned regions” are more extensive than footprint regions and cover footprint regions plus C and N- terminal protein regions.
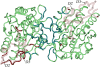
Figure 4. Disordered regions displayed in the homodimeric complex of heat shock protein hsp31 (1PV2).
The structured regions of 1PV2 are shown in green with interface residues highlighted in teal. A trace of possible order in the disordered regions was drawn using the 1ONS structure (the same protein in its monomeric state without disorder) which was structurally superimposed on 1PV2 two times, once for each half of the homodimer. Residues capping disordered regions in the homodimer are colored in red and the corresponding ordered residues from the monomer are drawn as silhouettes. A few additional residues of 1ONS in the left monomer are ordered compared to the same residues on the right and therefore are also colored red.
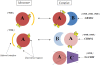
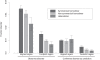
Figure 5. Average fraction disorder together with the standard error is plotted for three categories of oligomers: symmetrical, nonsymmetrical homodimers and heterodimers; for fraction disorder in the aligned and footprint regions.
The “footprint region” extends from the first to the last residue in the alignment mapping CDD family to a given chain. “Aligned regions” are more extensive than footprint regions and cover footprint regions plus C and N- terminal protein regions.
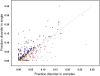
Figure 6. Fraction disorder in the alignment is plotted for monomeric and complex states of each protein averaged over families from “test588”.
Those cases with the disorder fraction in a monomer higher, lower or equal to the fraction disorder in a complex are shown with the circles, triangles and diamonds correspondingly. Entries from “test149” are shown as orange upside-down triangles.
Figure 7. Disorder-to-order transition upon complex formation in ubiquitin _C_-terminal hydrolase.
Structures of two forms of the hydrolase are shown to demonstrate the disordered region which becomes ordered upon complex formation. On the left side is the monomeric ubiquitin hydrolase (1UCH) with the residue at either end of the disordered region highlighted in red and shown with sidechains. On the right side is the complex between ubiquitin hydrolase and ubiquitin with the same residues highlighted in red and sidechains drawn. To trace the disordered region, the ordered region on the right has been mapped to the monomeric structure on the left using a superposition between the two structures and is shown as a silhouette.
Similar articles
- Analyzing molecular interactions.
Petsko GA. Petsko GA. Curr Protoc Bioinformatics. 2003 May;Chapter 8:Unit8.1. doi: 10.1002/0471250953.bi0801s01. Curr Protoc Bioinformatics. 2003. PMID: 18428708 Review. - Local structural disorder imparts plasticity on linear motifs.
Fuxreiter M, Tompa P, Simon I. Fuxreiter M, et al. Bioinformatics. 2007 Apr 15;23(8):950-6. doi: 10.1093/bioinformatics/btm035. Epub 2007 Mar 25. Bioinformatics. 2007. PMID: 17387114 - Interaction-site prediction for protein complexes: a critical assessment.
Zhou HX, Qin S. Zhou HX, et al. Bioinformatics. 2007 Sep 1;23(17):2203-9. doi: 10.1093/bioinformatics/btm323. Epub 2007 Jun 22. Bioinformatics. 2007. PMID: 17586545 Review. - Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein-protein interactions.
Guharoy M, Chakrabarti P. Guharoy M, et al. Bioinformatics. 2007 Aug 1;23(15):1909-18. doi: 10.1093/bioinformatics/btm274. Epub 2007 May 17. Bioinformatics. 2007. PMID: 17510165 - Protein-protein interactions.
Alexov E. Alexov E. Curr Pharm Biotechnol. 2008 Apr;9(2):55-6. doi: 10.2174/138920108783955182. Curr Pharm Biotechnol. 2008. PMID: 18393861 No abstract available.
Cited by
- Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins.
Zhao B, Ghadermarzi S, Kurgan L. Zhao B, et al. Comput Struct Biotechnol J. 2023 Jun 2;21:3248-3258. doi: 10.1016/j.csbj.2023.06.001. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 38213902 Free PMC article. - Conformational Heterogeneity and Frustration of the Tumor Suppressor p53 as Tuned by Punctual Mutations.
Bizzarri AR. Bizzarri AR. Int J Mol Sci. 2022 Oct 20;23(20):12636. doi: 10.3390/ijms232012636. Int J Mol Sci. 2022. PMID: 36293489 Free PMC article. - Modeling the Dynamics of Protein-Protein Interfaces, How and Why?
Karaca E, Prévost C, Sacquin-Mora S. Karaca E, et al. Molecules. 2022 Mar 11;27(6):1841. doi: 10.3390/molecules27061841. Molecules. 2022. PMID: 35335203 Free PMC article. Review. - Stabilization Effect of Intrinsically Disordered Regions on Multidomain Proteins: The Case of the Methyl-CpG Protein 2, MeCP2.
Ortega-Alarcon D, Claveria-Gimeno R, Vega S, Jorge-Torres OC, Esteller M, Abian O, Velazquez-Campoy A. Ortega-Alarcon D, et al. Biomolecules. 2021 Aug 16;11(8):1216. doi: 10.3390/biom11081216. Biomolecules. 2021. PMID: 34439881 Free PMC article. - PPIDomainMiner: Inferring domain-domain interactions from multiple sources of protein-protein interactions.
Alborzi SZ, Ahmed Nacer A, Najjar H, Ritchie DW, Devignes MD. Alborzi SZ, et al. PLoS Comput Biol. 2021 Aug 9;17(8):e1008844. doi: 10.1371/journal.pcbi.1008844. eCollection 2021 Aug. PLoS Comput Biol. 2021. PMID: 34370723 Free PMC article.
References
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. - PubMed
- Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. - PubMed
- Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. - PubMed
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. - PubMed
- Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24:325–342. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources