A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis - PubMed (original) (raw)
Case Reports
. 2009 Mar 13;323(5920):1473-7.
doi: 10.1126/science.1168979.
Marcella Catania, Michela Morbin, Giacomina Rossi, Silvia Suardi, Giulia Mazzoleni, Marco Merlin, Anna Rita Giovagnoli, Sara Prioni, Alessandra Erbetta, Chiara Falcone, Marco Gobbi, Laura Colombo, Antonio Bastone, Marten Beeg, Claudia Manzoni, Bruna Francescucci, Alberto Spagnoli, Laura Cantù, Elena Del Favero, Efrat Levy, Mario Salmona, Fabrizio Tagliavini
Affiliations
- PMID: 19286555
- PMCID: PMC2728497
- DOI: 10.1126/science.1168979
Case Reports
A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis
Giuseppe Di Fede et al. Science. 2009.
Abstract
beta-Amyloid precursor protein (APP) mutations cause familial Alzheimer's disease with nearly complete penetrance. We found an APP mutation [alanine-673-->valine-673 (A673V)] that causes disease only in the homozygous state, whereas heterozygous carriers were unaffected, consistent with a recessive Mendelian trait of inheritance. The A673V mutation affected APP processing, resulting in enhanced beta-amyloid (Abeta) production and formation of amyloid fibrils in vitro. Co-incubation of mutated and wild-type peptides conferred instability on Abeta aggregates and inhibited amyloidogenesis and neurotoxicity. The highly amyloidogenic effect of the A673V mutation in the homozygous state and its anti-amyloidogenic effect in the heterozygous state account for the autosomal recessive pattern of inheritance and have implications for genetic screening and the potential treatment of Alzheimer's disease.
Figures
Fig. 1
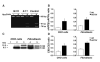
Analysis of APP gene and APP processing. (A) APP gene analysis by restriction fragment length polymorphism of 169—base pair (bp) polymerase chain reaction (PCR) products amplified from homozygous (III-16), heterozygous (II-11), and control subjects. In the absence of the A673V mutation, the enzyme HpyCH4V generates two fragments of 91 and 78 bp. The mutation abolishes the restriction site, and the PCR product remains uncut. (B) sAPPβ:sAPPα ratio in conditioned media from CHO cells transfected with wild-type or A673V-mutated APP and fibroblasts of the proband and four controls. Error bars represent means ± SD. (C) APP carboxy-terminal fragments C99 and C83 (arrowheads) in CHO cells transfected with wild-type (lane 2) or A673V-mutated (lane 3) APP and fibroblasts from a control (lane 4) and the proband (lane 5), as shown by immunoblot analysis. Lane 1 corresponds to from nontransfected CHO cells. (D) Densitometric analysis of immunoblots, showing a significant increase in the C99:C83 ratio (P = 0.0001) in cells carrying the A673V mutation. Error bars represent means ± SD.
Fig. 2
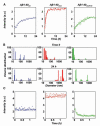
Short-time kinetics of Aβ assembly and disassembly, determined by laser light scattering. (A) Course of the light intensity scattered by solutions of Aβ1-40wt (blue), Aβ1-40mut (red), and their equimolar mixture (green). The corresponding exponential fits are indicated by full lines. (B) Particle size distribution of Aβ1-40wt (blue), Aβ1-40mut (red), and the peptide mixture (green) immediately after sample preparation (time 0) and after 24 hours. (C) Short-time dissolution kinetics of 48-hour-aged peptide aggregates after fivefold dilution with buffer. a.u., arbitrary units.
Fig. 3
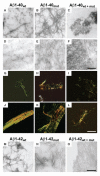
Aggregation properties of mutated and wild-type Aβ peptides. (A to F) Electron micrographs of aggregates generated by Aβ1-40wt, Aβ1-40mut, and equimolar mixtures after 72 hours [(A) to (C), negative staining] and 20 days incubation [(D) to (F), positive staining]. (G to L) Polarized light microscopy of Aβ aggregates stained with Congo red after 72 hours [(G) to (I)] and 20 days [(J) to (L)]. (M to O) Electron micrographs of negatively stained aggregates generated by Aβ1-42wt (M), Aβ1-42mut (N), and equimolar mixtures (O) after 5 days incubation. The peptide mixture contains mainly amorphous material (O), whereas wild-type and mutated Aβ1-42 are assembled in fibrillary structures. Scale bars indicate 250 nm [(A) to (F)], 50 μm [(G) to (L)], and 125 nm [(M) to (O)].
Fig. 4
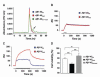
Physicochemical and biological properties of mutated and wild-type Aβ peptides. (A) Size exclusion chromatograms of Aβ1-40wt, Aβ1-40mut, and equimolar peptide mixture aggregates after treatment with 1 M urea for 24 hours. Monomeric species (arrow) are only seen in the peptide mixture. (B and C) Binding of wild-type and mutated Aβ1-40 (B) or Aβ1-6 (C) to amyloid fibrils of Aβ1-40wt determined by surface plasmon resonance. Solutions of Aβ1-40 (1 μM) or Aβ1-6 (500 μM) were injected onto Aβ1-40wt fibrils immobilized on the sensor chip for the time indicated by the bars. (D) Viability of human neuroblastoma cells after 24 hours exposure to 5 μM Aβ1-42wt, Aβ1-42mut, and the equimolar mixture. Error bars represent SD of the mean of eight replicates. *P = 0.026, **P < 0.001.
Similar articles
- Good gene, bad gene: new APP variant may be both.
Di Fede G, Catania M, Morbin M, Giaccone G, Moro ML, Ghidoni R, Colombo L, Messa M, Cagnotto A, Romeo M, Stravalaci M, Diomede L, Gobbi M, Salmona M, Tagliavini F. Di Fede G, et al. Prog Neurobiol. 2012 Dec;99(3):281-92. doi: 10.1016/j.pneurobio.2012.06.004. Epub 2012 Jun 19. Prog Neurobiol. 2012. PMID: 22727994 Review. - Neuropathology of the recessive A673V APP mutation: Alzheimer disease with distinctive features.
Giaccone G, Morbin M, Moda F, Botta M, Mazzoleni G, Uggetti A, Catania M, Moro ML, Redaelli V, Spagnoli A, Rossi RS, Salmona M, Di Fede G, Tagliavini F. Giaccone G, et al. Acta Neuropathol. 2010 Dec;120(6):803-12. doi: 10.1007/s00401-010-0747-1. Epub 2010 Sep 15. Acta Neuropathol. 2010. PMID: 20842367 - APP Osaka Mutation in Familial Alzheimer's Disease-Its Discovery, Phenotypes, and Mechanism of Recessive Inheritance.
Tomiyama T, Shimada H. Tomiyama T, et al. Int J Mol Sci. 2020 Feb 19;21(4):1413. doi: 10.3390/ijms21041413. Int J Mol Sci. 2020. PMID: 32093100 Free PMC article. Review. - BACE1 Cleavage Site Selection Critical for Amyloidogenesis and Alzheimer's Pathogenesis.
Zhang S, Wang Z, Cai F, Zhang M, Wu Y, Zhang J, Song W. Zhang S, et al. J Neurosci. 2017 Jul 19;37(29):6915-6925. doi: 10.1523/JNEUROSCI.0340-17.2017. Epub 2017 Jun 16. J Neurosci. 2017. PMID: 28626014 Free PMC article. - Alternative Selection of β-Site APP-Cleaving Enzyme 1 (BACE1) Cleavage Sites in Amyloid β-Protein Precursor (APP) Harboring Protective and Pathogenic Mutations within the Aβ Sequence.
Kimura A, Hata S, Suzuki T. Kimura A, et al. J Biol Chem. 2016 Nov 11;291(46):24041-24053. doi: 10.1074/jbc.M116.744722. Epub 2016 Sep 29. J Biol Chem. 2016. PMID: 27687728 Free PMC article.
Cited by
- Association of APP gene polymorphisms and promoter methylation with essential hypertension in Guizhou: a case-control study.
Li R, Song J, Zhao A, Diao X, Zhang T, Qi X, Guan Z, An Y, Ren L, Wang C, He Y. Li R, et al. Hum Genomics. 2023 Mar 20;17(1):25. doi: 10.1186/s40246-023-00462-y. Hum Genomics. 2023. PMID: 36941702 Free PMC article. - Attempt to Untangle the Prion-Like Misfolding Mechanism for Neurodegenerative Diseases.
Sarnataro D. Sarnataro D. Int J Mol Sci. 2018 Oct 9;19(10):3081. doi: 10.3390/ijms19103081. Int J Mol Sci. 2018. PMID: 30304819 Free PMC article. Review. - Amyloid Precursor Protein A713T Mutation in Calabrian Patients with Alzheimer's Disease: A Population Genomics Approach to Estimate Inheritance from a Common Ancestor.
Abondio P, Sarno S, Giuliani C, Laganà V, Maletta R, Bernardi L, Bruno F, Colao R, Puccio G, Frangipane F, Borroni B, Van Broeckhoven C, Luiselli D, Bruni A. Abondio P, et al. Biomedicines. 2021 Dec 23;10(1):20. doi: 10.3390/biomedicines10010020. Biomedicines. 2021. PMID: 35052700 Free PMC article. - Brain transcriptomes of zebrafish and mouse Alzheimer's disease knock-in models imply early disrupted energy metabolism.
Barthelson K, Newman M, Lardelli M. Barthelson K, et al. Dis Model Mech. 2022 Jan 1;15(1):dmm049187. doi: 10.1242/dmm.049187. Epub 2022 Jan 26. Dis Model Mech. 2022. PMID: 34842276 Free PMC article. - Effect of the English familial disease mutation (H6R) on the monomers and dimers of Aβ40 and Aβ42.
Viet MH, Nguyen PH, Derreumaux P, Li MS. Viet MH, et al. ACS Chem Neurosci. 2014 Aug 20;5(8):646-57. doi: 10.1021/cn500007j. Epub 2014 Jun 30. ACS Chem Neurosci. 2014. PMID: 24949887 Free PMC article.
References
- Hardy J, Selkoe DJ. Science. 2002;297:353. - PubMed
- Selkoe DJ. Physiol. Rev. 2001;81:741. - PubMed
- Walsh DM, et al. Nature. 2002;416:535. - PubMed
- Rocchi A, Pellegrini S, Siciliano G, Murri L. Brain Res. Bull. 2003;61:1. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases