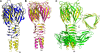Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages - PubMed (original) (raw)
Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages
Marina Siponen et al. J Bacteriol. 2009 May.
Abstract
Lactococcus lactis, a gram-positive bacterium widely used by the dairy industry to manufacture cheeses, is subject to infection by a diverse population of virulent phages. We have previously determined the structures of three receptor binding proteins (RBPs) from lactococcal phages TP901-1, p2, and bIL170, each of them having a distinct host range. Virulent phages p2 and bIL170 are classified within the 936 group, while the temperate phage TP901-1 is a member of the genetically distinct P335 polythetic group. These RBPs comprise three domains: the N-terminal domain, binding to the virion particle; a beta-helical linker domain; and the C-terminal domain, bearing the receptor binding site used for host recognition. Here, we have designed, expressed, and determined the structure of an RBP chimera in which the N-terminal and linker RBP domains of phage TP901-1 (P335) are fused to the C-terminal RBP domain of phage p2 (936). This chimera exhibits a stable structure that closely resembles the parental structures, while a slight displacement of the linker made RBP domain adaptation efficient. The receptor binding site is structurally indistinguishable from that of native p2 RBP and binds glycerol with excellent affinity.
Figures
FIG. 1.
Structures and sequences of RBPs from lactococcal phages. (A) Three-dimensional structure of the RBP from phage TP901-1 (P335 group; blue). (B) Three-dimensional structure of the RBP from phage p2 (936 group; magenta). (C) View of a model associating domains of TP901-1 (N terminus and linker domain, below red line, blue) and p2 (head, above red line, magenta) RBPs. (D) Three-dimensional crystal structure of chimera form 1 (yellow) assembled according to the model in panel C. (E) Sequence alignment of the RBPs of p2 (part) and TP901-1. The secondary structure is described above the alignment. The binding residues are shown with blue dots. The hinge proline (Pro 162/63) is identified by a red arrow. The chimera is composed of the N-terminal domain (residues 17 to 33) and the linker domain residues (residues 34 to 63) from phage TP901-1 RBP and the C-terminal domain (residues 163 to 264) from phage p2 RBP.
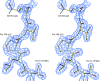
FIG. 2.
Superimposition and comparison of the RBPs from lactococcal phages p2 and TP901-1 as well as their chimera. (A) Superimposition of chimera form 1 (yellow) and form 2 (blue). (B) Superimposition, using the N-terminal and linker domains, of form 2 (yellow) on the wild-type RBP of phage TP901-1 (pink). (C) Superimposition, using the C-terminal domains, of form 2 (yellow) on the wild-type RBP of phage p2 (green). Inset, 90° view of the β-prism linker domains of p2 (green) and the chimera (yellow), illustrating the larger size of the latter.
FIG. 3.
2fo-fc 1.65-Å resolution electron density (stereo view) contoured at 1σ of the stretch of residues linking the swapped domains of the chimera, including the hinge proline.
FIG. 4.
Identification of the protease cleavage sites on RBPs and their correlation with domain junctions. (A) Chimera form 1 and its cleavage after Glu16. (B) Chimera form 2 and its cleavage after Thr30, between the N terminus and linker domains. (C) RBP head domain of phage p2 from the complex with llama VHHs.
Similar articles
- Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages.
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Dunne M, et al. Viruses. 2018 Jul 28;10(8):397. doi: 10.3390/v10080397. Viruses. 2018. PMID: 30060549 Free PMC article. Review. - Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein.
Farenc C, Spinelli S, Vinogradov E, Tremblay D, Blangy S, Sadovskaya I, Moineau S, Cambillau C. Farenc C, et al. J Virol. 2014 Jun;88(12):7005-15. doi: 10.1128/JVI.00739-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719416 Free PMC article. - Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1.
Spinelli S, Campanacci V, Blangy S, Moineau S, Tegoni M, Cambillau C. Spinelli S, et al. J Biol Chem. 2006 May 19;281(20):14256-62. doi: 10.1074/jbc.M600666200. Epub 2006 Mar 20. J Biol Chem. 2006. PMID: 16549427 - The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. Legrand P, et al. mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article. - Host recognition by lactic acid bacterial phages.
Mahony J, Cambillau C, van Sinderen D. Mahony J, et al. FEMS Microbiol Rev. 2017 Aug 1;41(Supp_1):S16-S26. doi: 10.1093/femsre/fux019. FEMS Microbiol Rev. 2017. PMID: 28830088 Review.
Cited by
- Structures and host-adhesion mechanisms of lactococcal siphophages.
Spinelli S, Veesler D, Bebeacua C, Cambillau C. Spinelli S, et al. Front Microbiol. 2014 Jan 16;5:3. doi: 10.3389/fmicb.2014.00003. eCollection 2014. Front Microbiol. 2014. PMID: 24474948 Free PMC article. Review. - Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages.
Kot W, Hammer K, Neve H, Vogensen FK. Kot W, et al. Appl Environ Microbiol. 2013 May;79(10):3311-4. doi: 10.1128/AEM.00012-13. Epub 2013 Mar 15. Appl Environ Microbiol. 2013. PMID: 23503306 Free PMC article. - Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages.
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Dunne M, et al. Viruses. 2018 Jul 28;10(8):397. doi: 10.3390/v10080397. Viruses. 2018. PMID: 30060549 Free PMC article. Review. - Progress in lactic acid bacterial phage research.
Mahony J, Bottacini F, van Sinderen D, Fitzgerald GF. Mahony J, et al. Microb Cell Fact. 2014 Aug 29;13 Suppl 1(Suppl 1):S1. doi: 10.1186/1475-2859-13-S1-S1. Epub 2014 Aug 29. Microb Cell Fact. 2014. PMID: 25185514 Free PMC article. Review. - Bacteriophage-host interactions as a platform to establish the role of phages in modulating the microbial composition of fermented foods.
White K, Yu JH, Eraclio G, Dal Bello F, Nauta A, Mahony J, van Sinderen D. White K, et al. Microbiome Res Rep. 2022 Jan 12;1(1):3. doi: 10.20517/mrr.2021.04. eCollection 2022. Microbiome Res Rep. 2022. PMID: 38089066 Free PMC article. Review.
References
- Ackermann, H. W. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152227-243. - PubMed
- Argos, P., T. Tsukihara, and M. G. Rossmann. 1980. A structural comparison of concanavalin A and tomato bushy stunt virus protein. J. Mol. Evol. 15169-179. - PubMed
- Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16673-685. - PubMed
- Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98825-833. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources