The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells - PubMed (original) (raw)
. 2009 Apr;119(4):826-36.
doi: 10.1172/JCI36428. Epub 2009 Mar 16.
Affiliations
- PMID: 19287094
- PMCID: PMC2662549
- DOI: 10.1172/JCI36428
The TEL-AML1 leukemia fusion gene dysregulates the TGF-beta pathway in early B lineage progenitor cells
Anthony M Ford et al. J Clin Invest. 2009 Apr.
Abstract
Chromosome translocation to generate the TEL-AML1 (also known as ETV6-RUNX1) chimeric fusion gene is a frequent and early or initiating event in childhood acute lymphoblastic leukemia (ALL). Our starting hypothesis was that the TEL-AML1 protein generates and maintains preleukemic clones and that conversion to overt disease requires secondary genetic changes, possibly in the context of abnormal immune responses. Here, we show that a murine B cell progenitor cell line expressing inducible TEL-AML1 proliferates at a slower rate than parent cells but is more resistant to further inhibition of proliferation by TGF-beta. This facilitates the competitive expansion of TEL-AML1-expressing cells in the presence of TGF-beta. Further analysis indicated that TEL-AML1 binds to a principal TGF-beta signaling target, Smad3, and compromises its ability to activate target promoters. In mice expressing a TEL-AML1 transgene, early, pre-pro-B cells were increased in number and also showed reduced sensitivity to TGF-beta-mediated inhibition of proliferation. Moreover, expression of TEL-AML1 in human cord blood progenitor cells led to the expansion of a candidate preleukemic stem cell population that had an early B lineage phenotype (CD34+CD38-CD19+) and a marked growth advantage in the presence of TGF-beta. Collectively, these data suggest a plausible mechanism by which dysregulated immune responses to infection might promote the malignant evolution of TEL-AML1-expressing preleukemic clones.
Figures
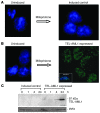
Figure 1. Induction of TEL-AML1 protein expression in BaF3 cells.
(A) Uninduced (left) and mifepristone-induced control cells (right) stained with both DAPI and an antibody against the TEL-AML1 V5 tag (original magnification, ×100). (B) Uninduced (left, DAPI and anti–V5 tag stained) and mifepristone-induced expression of TEL-AML1 (right, anti–V5 tag alone) (original magnification, ×100). The inset (×100) shows a confocal microscopy cross section. For DAPI staining, see Supplemental Figure 2E. (C) Western blot analysis over time of mifepristone-induced control and TEL-AML1–expressing BaF3 cell whole-cell protein extracts, blotted with anti-AML1 antibody. A loading control (IFN regulatory factor 2 [IRF2]) is shown below.
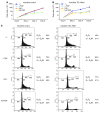
Figure 2. Effects of TEL-AML1 expression and TGF-β on growth rates and cell cycle of BaF3 cells.
(A) Typical growth curve of control cells grown in the presence or absence of mifepristone inducer (Ind) and/or TGF-β (TGF) at 10 ng/ml. (B) Typical growth curve of clone 1/27 cells (inducible for TEL-AML1) grown in the presence or absence of inducer and/or TGF-β at 10 ng/ml. (C) Cell cycle analysis (BrdU) of inducible control and inducible TEL-AML1 cells grown in the presence or absence of inducer and/or TGF-β.
Figure 3. Competition assay of a mix of TEL-AML1–expressing and –nonexpressing BaF3 cells grown in the presence or absence of TGF-β.
A mixture of TEL-AML1–expressing and –nonexpressing cells (84%:16%) was grown in the presence or absence of TGF-β, and the cell growth and TEL-AML1 expression profile were analyzed over time. Growth of TEL-AML1–expressing cells but not control cells is sustained in the presence of TGF-β. Flow cytometry analysis: the vertical axis represents fluorescence activity (for TEL-AML1 expression using an antibody against the V5 tag); the horizontal axis represents light scatter (cell size). In 4 repeat experiments, the starting percentage of TEL-AML1–positive cells was around 80%, and their persistence in the presence of TGF-β was consistent.
Figure 4. TEL-AML1 activates expression of p21 but blocks the TGF-β–mediated activation of p27 and inhibits the TGF-β response of a target gene promoter.
(A) Q-PCR analysis showing activation of p21 (CDKN1A) by TEL-AML1 in the absence of TGF-β. Control cells and cells inducible for TEL-AML1 were incubated for 3 days in the absence (–) or presence of inducer. cDNA was subjected to Q-PCR and normalized to GAPDH, to which relative expression of p21 is shown. Error bars represent the SD of an experiment performed in triplicate and repeated 3 times. (B) Q-PCR analysis showing a block in expression of TGF-β–induced p27KIP1 in the presence of TEL-AML1. cDNA was prepared from a TGF-β time-course analysis of both control and TEL-AML1–inducible cells in the presence or absence of the TEL-AML1–inducing agent (ind). Cells inducible for TEL-AML1 but not actually induced are indicated by parentheses, i.e., (TEL-AML1). Experiments were repeated 3 times. (C) Inhibition of the TGF-β–responsive IgA promoter by TEL-AML1 in a luciferase reporter assay. Activation of the IgA promoter was assayed by its transient transfection into control cells and cells inducible for TEL-AML1. Cells inducible for TEL-AML1 but not actually induced are indicated by parentheses. Growth was continued in the presence of TGF-β, either alone or after addition of the TEL-AML1–inducing agent. Error bars represent SD from 3 independent experiments. (D) TEL-AML1 associates with Smad3. Cell lysates from control and TEL-AML1–expressing cells were immunoprecipitated with anti-Smad3 antibody and half the IP subjected to Western blot analysis with an antibody against the runt homology domain (RHD) of AML1. Lane 1, uninduced cells; lane 2, TEL-AML1–induced; lane 3, TEL-AML1–induced + TGF; lane 4, control cells + TGF.
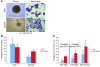
Figure 5. Impact of TEL-AML1 expression on B progenitor CFC numbers and their inhibition by TGF-β.
(A) Typical first-round colony morphology in a B progenitor cell colony assay. Colonies (×40) were grown in the presence of IL-7 with additional SCF and Flt-3 ligand, picked after 9 days growth, spread on glass slides, and stained with Giemsa for morphology (×100). (B) First-plate phenotype. Average colony numbers from first plating of BM cells isolated from wild-type and TEL-AML1 transgenic lines. Cells (7 × 105) were plated in methylcellulose for 9 days under B cell growth conditions with additional SCF and Flt-3 ligand. Error bars represent SD from 5 independent experiments. (C) Average resistance to TGF-β. Effect of TGF-β on second-round colonies picked from first-round tight colonies (which yield only tight colonies) and spread colonies (which yield both spread and tight colonies). Average resistance was calculated as the number of colonies in the presence of TGF-β divided by the total number of colonies in its absence in the same experiment. Error bars represent SD from 5 independent experiments.
Figure 6. Colony phenotypes from Eμ TEL-AML1–transgenic lines and wild-type controls.
Analysis of cell immunophenotype by flow cytometry. Tight and spread colonies isolated from wild-type and TEL-AML1–transgenic lines were compared by surface phenotype using monoclonal antibodies against B220, CD19, Flt-3, BP1 (63), c-Kit, Sca-1, IL-7R, and CD11b, as well as isotype controls. The percentage of cells within each quadrant is shown in gray, and the percentage within a specific gate is shown in red.
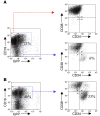
Figure 7. TGF-β “selects” candidate human pre-LSCs.
Cord blood stem cell encoded (CD34+CD38–) populations were transduced with lentiviral TEL-AML1 and plated on MS-5 stroma for 3 weeks to generate pre-LSCs (CD34+CD38–CD19+) (13). Cells were replated on MS-5 with or without TGF-β. (A) Cell populations without TGF-β. (B) Cell populations with TGF-β. The percentage of cells in each particular gate or quadrant is shown from 1 experiment but was replicated and consistent in 2 separate experiments (i.e., 3 in total).
Similar articles
- The TEL-AML1 fusion protein of acute lymphoblastic leukemia modulates IRF3 activity during early B-cell differentiation.
de Laurentiis A, Hiscott J, Alcalay M. de Laurentiis A, et al. Oncogene. 2015 Dec 3;34(49):6018-28. doi: 10.1038/onc.2015.50. Epub 2015 Apr 20. Oncogene. 2015. PMID: 25893288 - Modeling first-hit functions of the t(12;21) TEL-AML1 translocation in mice.
Tsuzuki S, Seto M, Greaves M, Enver T. Tsuzuki S, et al. Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8443-8. doi: 10.1073/pnas.0402063101. Epub 2004 May 20. Proc Natl Acad Sci U S A. 2004. PMID: 15155899 Free PMC article. - The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.
Lo Coco F, Pisegna S, Diverio D. Lo Coco F, et al. Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review. - Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia.
Zelent A, Greaves M, Enver T. Zelent A, et al. Oncogene. 2004 May 24;23(24):4275-83. doi: 10.1038/sj.onc.1207672. Oncogene. 2004. PMID: 15156184 Review.
Cited by
- Identification of Gene Regulatory Networks in B-Cell Progenitor Differentiation and Leukemia.
Nagel S, Meyer C. Nagel S, et al. Genes (Basel). 2024 Jul 24;15(8):978. doi: 10.3390/genes15080978. Genes (Basel). 2024. PMID: 39202339 Free PMC article. - The Diverse Roles of ETV6 Alterations in B-Lymphoblastic Leukemia and Other Hematopoietic Cancers.
Monovich AC, Gurumurthy A, Ryan RJH. Monovich AC, et al. Adv Exp Med Biol. 2024;1459:291-320. doi: 10.1007/978-3-031-62731-6_13. Adv Exp Med Biol. 2024. PMID: 39017849 Review. - Impact of genetic alterations on central nervous system progression of primary vitreoretinal lymphoma.
Yoshifuji K, Sadato D, Toya T, Motomura Y, Hirama C, Takase H, Yamamoto K, Harada Y, Mori T, Nagao T. Yoshifuji K, et al. Haematologica. 2024 Nov 1;109(11):3641-3649. doi: 10.3324/haematol.2023.284953. Haematologica. 2024. PMID: 38841798 Free PMC article. - Inflammation as a driver of hematological malignancies.
Saluja S, Bansal I, Bhardwaj R, Beg MS, Palanichamy JK. Saluja S, et al. Front Oncol. 2024 Mar 20;14:1347402. doi: 10.3389/fonc.2024.1347402. eCollection 2024. Front Oncol. 2024. PMID: 38571491 Free PMC article. Review. - SPAG6 regulates cell proliferation and apoptosis via TGF-β/Smad signal pathway in adult B-cell acute lymphoblastic leukemia.
Zhao B, Yin J, Ding L, Luo J, Luo J, Mu J, Pan S, Du J, Zhong Y, Zhang L, Liu L. Zhao B, et al. Int J Hematol. 2024 Feb;119(2):119-129. doi: 10.1007/s12185-023-03684-x. Epub 2023 Dec 26. Int J Hematol. 2024. PMID: 38147275
References
- Romana S.P., et al. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. - PubMed
- Shurtleff S.A., et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. . Leukemia. 1995;9:1985–1989. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials