Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae - PubMed (original) (raw)
Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae
Moumita Biswas et al. Mol Cell Biol. 2009 May.
Abstract
In Saccharomyces cerevisiae, transcriptional silencing occurs at the cryptic mating-type loci (HML and HMR), telomeres, and ribosomal DNA (rDNA; RDN1). Silencing in the rDNA is unusual in that polymerase II (Pol II) promoters within RDN1 are repressed by Sir2 but not Sir3 or Sir4. rDNA silencing unidirectionally spreads leftward, but the mechanism of limiting its spreading is unclear. We searched for silencing barriers flanking the left end of RDN1 by using an established assay for detecting barriers to HMR silencing. Unexpectedly, the unique sequence immediately adjacent to RDN1, which overlaps a prominent cohesin binding site (CARL2), did not have appreciable barrier activity. Instead, a fragment located 2.4 kb to the left, containing a tRNA(Gln) gene and the Ty1 long terminal repeat, had robust barrier activity. The barrier activity was dependent on Pol III transcription of tRNA(Gln), the cohesin protein Smc1, and the SAS1 and Gcn5 histone acetyltransferases. The location of the barrier correlates with the detectable limit of rDNA silencing when SIR2 is overexpressed, where it blocks the spreading of rDNA heterochromatin. We propose a model in which normal Sir2 activity results in termination of silencing near the physical rDNA boundary, while tRNA(Gln) blocks silencing from spreading too far when nucleolar Sir2 pools become elevated.
Figures
FIG. 1.
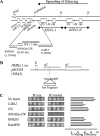
The tRNAGln-Ty1 LTR from chromosome XII is a strong barrier to silencing. (A) Map of chromosome XII showing the RDN1 locus and the positions of the test fragments used in the barrier assay. The arrow indicates the direction of RNA Pol I-driven transcription and spreading of silencing. (B) Principle of the mating assay used to measure barrier activity (14). (C) Results from a quantitative mating experiment showing that the DNA fragment containing the tRNAGln and LTR acts as a strong barrier. The 10-fold serial dilutions of the test culture shown range from 1 (i.e., undiluted) to 0.001 (i.e., 1,000-fold diluted). A quantitative mating assay (described in Materials and Methods) was used to assess barrier activity. The vector pRO363 with no insert or a 1.4-kb bacterial gene for Geneticin resistance, KanMX6 (which does not have barrier activity and behaves as a stuffer fragment in this assay), was used as a mating control. The SGD coordinates of the fragments tested are as follows: CARL2, 450143 to 451532; IVS, 449029 to 450203; the tRNAGln and the LTR, 448623 to 449050; RNH203, 447359 to 448488. A quantitative estimate of the mating efficiency of each of the fragments is depicted in the histogram.
FIG. 2.
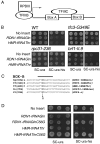
RNA Pol III-driven transcription and tDNA barrier activity. (A) Schematic showing the organization of the internal promoters of tRNA genes. The rectangular boxes represent the conserved box A and box B ICRs. The arrow indicates the start site of tRNA transcription. (B) The barrier activity of tRNAGln is mildly reduced in RNA Pol III complex mutants. The barrier activity of the tRNAGln and _HMR_-tRNAThr was tested in the wild type (WT) and mutants defective in various RNA Pol III complex subunits (_tfc3G349E, rpc31_-236, and _brf1_-II.6 mutants). The barrier test constructs were maintained episomally in these strains; the results shown for the _brf1_-II.9 mutant were obtained by integration of the barrier test constructs. (C) Multiple sequence alignment of box B ICRs of _RDN1_-tRNAGln with four other tDNAs whose transcription (_SUP53_-tRNALeu and _SUP4_-o) and barrier activity (_HMR_-tRNAThr, _TRT2_-tRNAThr) requirements have been previously analyzed. Sequences flanking the ICR are also shown. The solid black line indicates the positions of the box B ICRs. C56 residues that are known to be required for transcription or that were tested for barrier activity are shown in bold. The arrow indicates the position of the box B mutation. (D) Effect of C56G box B mutation on barrier activity. Representative results from a mating assay showing that the barrier activity of tRNAGln is abrogated by the C56G promoter mutation. The wild-type (pRO466) and C56G mutant (pRO468) _HMR_-tRNAThr were also tested in parallel as a positive control. Each row shows spots derived from mating assays done with various amounts of cells (10×, 1×, and 0.1×) of the same test culture.
FIG. 3.
HATs are required for tDNA barrier activity. The barrier activity of _RDN1_-tRNAGln and _HMR_-tRNAThr was measured in sas2, sas4, sas5, gcn5, sas3, and esa1 HAT mutants by a quantitative mating assay. The two spots shown represent two different dilutions of the same culture. Diploids of the sas3 and sas5 mutants were selected on SC-Ura-Lys plates. WT, wild type.
FIG. 4.
Cohesin dependence of tDNA barrier activity. The barrier activity of _RDN1_-tRNAGln and _HMR_-tRNAThr is affected in an _smc1_-2 mutant. Three or more transformants corresponding to each tDNA were tested for barrier activity in wild-type (WT) and _smc1_-2 mutant cells by a quantitative mating assay. Diploids were selected on SC-Ura plates. Each row shows spots derived from mating assays done with various amounts (10×, 1×, and 0.1×) of cells from the same test culture. The panels on the right show the temperature-sensitive phenotype of the _smc1_-2 mutant Ura+ transformants. Representative results are shown.
FIG. 5.
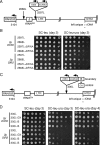
The position of a tRNAGln gene correlates with the spreading limit for rDNA silencing. (A) Schematic diagram showing various positions at which the _mURA3_-HIS3 reporter gene cassette was integrated into the unique chromosome XII sequence flanking the centromere-proximal (left) end of the rDNA tandem array, which is defined by the NTS1 sequence. Vertical arrows indicate the positions of insertions (not to scale). The number of nucleotides to the left of the NTS1 rDNA sequence are indicated by 50L, 300L, etc. The tRNAGln gene is located at positions 2697L and 2769L (SGD coordinate 448722). The next closest gene is RNH203, the open reading frame of which begins 3,104 bp to the left of the rDNA (SGD coordinate 448315). (B) Silencing is normally not detected near the tRNAGln gene. TRP1 indicates a strain in which _mURA3_-HIS3 was integrated at the trp1_Δ_63 locus and is not silenced. Silencing is indicated by poor growth on the SC-Ura plate. (C) Effects of a high-copy 2μm SIR2 plasmid (pSB766) on silencing at positions surrounding the tRNA gene. The empty vector is pRS425. (D) Effect of high-copy SIR2 on a strain in which the _mURA3_-HIS3 reporter precisely replaces the tRNA gene, inserting the mURA3 gene at position 2697L.
FIG. 6.
The tRNAGln gene blocks the spreading of rDNA silencing. (A) Schematic diagram showing the _mURA3_-HIS3 cassette integrated at the 2597L (SGD: XII-448822) or 2868L (SGD: XII-448551) position flanking the left end of the rDNA array. Where indicated, the tRNAGln gene was deleted (Δ). (B) SIR2 overexpression modestly induces silencing at the 2868L position if tRNAGln is deleted from chromosome XII (ΔtRNA). (C) A 427-bp boundary element fragment, B, containing the tRNAGln gene was ligated next to the mURA3 promoter and then integrated at the 300L position. Alternatively, a 427-bp fragment of φX174 DNA was used as a control stuffer fragment and also integrated at 300L. (D) The 427-bp boundary element fragment, but not the φX174 control fragment, partially blocked the spread of silencing into the 300L position when SIR2 was overexpressed.
Similar articles
- Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p.
Smith JS, Brachmann CB, Pillus L, Boeke JD. Smith JS, et al. Genetics. 1998 Jul;149(3):1205-19. doi: 10.1093/genetics/149.3.1205. Genetics. 1998. PMID: 9649515 Free PMC article. - Reconstitution of heterochromatin-dependent transcriptional gene silencing.
Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock CL, Moazed D. Johnson A, et al. Mol Cell. 2009 Sep 24;35(6):769-81. doi: 10.1016/j.molcel.2009.07.030. Mol Cell. 2009. PMID: 19782027 Free PMC article. - Isw1 acts independently of the Isw1a and Isw1b complexes in regulating transcriptional silencing at the ribosomal DNA locus in Saccharomyces cerevisiae.
Mueller JE, Bryk M. Mueller JE, et al. J Mol Biol. 2007 Aug 3;371(1):1-10. doi: 10.1016/j.jmb.2007.04.089. Epub 2007 May 18. J Mol Biol. 2007. PMID: 17561109 Free PMC article. - A model for step-wise assembly of heterochromatin in yeast.
Moazed D, Rudner AD, Huang J, Hoppe GJ, Tanny JC. Moazed D, et al. Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review. - The Sir2 family of protein deacetylases.
Blander G, Guarente L. Blander G, et al. Annu Rev Biochem. 2004;73:417-35. doi: 10.1146/annurev.biochem.73.011303.073651. Annu Rev Biochem. 2004. PMID: 15189148 Review.
Cited by
- RNA Polymerase I and Fob1 contributions to transcriptional silencing at the yeast rDNA locus.
Buck SW, Maqani N, Matecic M, Hontz RD, Fine RD, Li M, Smith JS. Buck SW, et al. Nucleic Acids Res. 2016 Jul 27;44(13):6173-84. doi: 10.1093/nar/gkw212. Epub 2016 Apr 8. Nucleic Acids Res. 2016. PMID: 27060141 Free PMC article. - Cohesin dysfunction results in cell wall defects in budding yeast.
Kothiwal D, Gopinath S, Laloraya S. Kothiwal D, et al. Genetics. 2021 Mar 3;217(1):1-16. doi: 10.1093/genetics/iyaa023. Genetics. 2021. PMID: 33683362 Free PMC article. - A positive but complex association between meiotic double-strand break hotspots and open chromatin in Saccharomyces cerevisiae.
Berchowitz LE, Hanlon SE, Lieb JD, Copenhaver GP. Berchowitz LE, et al. Genome Res. 2009 Dec;19(12):2245-57. doi: 10.1101/gr.096297.109. Epub 2009 Oct 2. Genome Res. 2009. PMID: 19801530 Free PMC article. - Genome-wide analysis of functional sirtuin chromatin targets in yeast.
Li M, Valsakumar V, Poorey K, Bekiranov S, Smith JS. Li M, et al. Genome Biol. 2013 May 27;14(5):R48. doi: 10.1186/gb-2013-14-5-r48. Genome Biol. 2013. PMID: 23710766 Free PMC article. - tRNAs as a Driving Force of Genome Evolution in Yeast.
Guimarães AR, Correia I, Sousa I, Oliveira C, Moura G, Bezerra AR, Santos MAS. Guimarães AR, et al. Front Microbiol. 2021 Mar 11;12:634004. doi: 10.3389/fmicb.2021.634004. eCollection 2021. Front Microbiol. 2021. PMID: 33776966 Free PMC article. Review.
References
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. Greene Publishing Associates, New York, NY.
- Bi, X., and J. R. Broach. 2001. Chromatin boundaries in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 11199-204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases