Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse - PubMed (original) (raw)
Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse
Enyuan Shang et al. Dev Dyn. 2009 Apr.
Abstract
The BET subfamily of bromodomain-containing genes is characterized by the presence of two bromodomains and a unique ET domain at their carboxyl termini. Here, we show that the founding member of this subfamily, Brd2, is an essential gene by generating a mutant mouse line lacking Brd2 function. Homozygous Brd2 mutants are embryonic lethal, with most Brd2(-/-) embryos dying by embryonic day 11.5. Before death, the homozygous embryos were notably smaller and exhibited abnormalities in the neural tube where the gene is highly expressed. Brd2-deficient embryonic fibroblast cells were observed to proliferate more slowly than controls. Experiments to explore whether placental insufficiency could be a cause of the embryonic lethality showed that injecting diploid mutant embryonic stem cells into tetraploid wild-type blastocysts did not rescue the lethality; that is Brd2-deficient embryos could not be rescued by wild-type extraembryonic tissues. Furthermore, there were enhanced levels of cell death in Brd2-deficient embryos.
Copyright 2009 Wiley-Liss, Inc.
Figures
Figure 1
Panel A: Schematic depiction of the inactivation of the Brd2 gene by a gene-trap approach. The gene trap vector was inserted into the first intron of the Brd2 gene, near the putative translation start codon ATG. The gene trap insertion is about 8 kb in size and is not depicted in proportion to the Brd2 gene. Arrows indicate the position of PCR primers used for verification of the insertion event and for genotyping progeny carrying the mutant or wild-type alleles. The primers are: EX1F (exon 1 forward), IN1R (intron 1 reverse), and GT1R (genetrap vector reverse). Panel B: Visualization of the PCR products used to genotype the progeny. The use of EX1F + GT1R gives the higher band (500 bp) representing the mutated allele; EX1F + IN1R produces the lower band (220 bp)--wildtype allele. Since the insertion is 8 kb, under our PCR condition (1 min extension time), this pair of primers would not yield a band corresponding to the mutant allele.
Figure 2
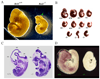
Panel A: Photographs of wild-type (Brd2+/+, left) and _Brd2_−/− (right) embryos at E9.5 of gestation. The mutant embryo is smaller and the developing neural tube is unfused (arrow). Panel B: A litter of 11 embryos at E9.5 of gestation were fixed in 4% paraformaldehyde and photographed together to portray the variations in phenotype among the embryos. The genotype and weight of the embryos are, from the top row and left to right: Top Row: +/+, 4.6 mg; +/+, 6.5 mg; +/−, 4.3 mg; +/−, 2.3 mg; Middle Row: +/−, 1.9 mg; +/+, 2.0 mg; −/−, 2.2 mg; +/−, 1.9 mg; and Bottom Row: −/−, 1.1 mg; −/−, 1.0 mg; −/−, 1.2 mg. The mean and SD for each genotype are: +/+, 5.1 ± 1.2 mg; +/−, 2.6 ± 1.3 mg; −/−, 1.4 ± 0.6 mg. Panel C: Sagittal sections of E11.5 embryos of wildtype and mutant embryos. Grossly normal appearing heart, lung, etc. are seen in the mutant, but the brain of the mutant is aberrant. FB, forebrain; MB, midbrain; HB, hindbrain. Panel D: Whole-mount in situ hybridization of wild-type and Brd2 mutant E9.5 embryos using digoxygenin-labeled Brd2 RNA probes (see Materials and Methods). Brd2 mRNA is detected in the wild-type developing neural system but is absent in the mutant.
Figure 3
Panel A: Growth parameters of mouse embryonic fibroblast (MEFs) derived from Brd2+/+, Brd2+/−, and _Brd2_−/− embryos of E9.5 (157-5 and 157-11), E10.5 (128-1 and 128-2), and E11.5 (127-2 and 127-7). The cells were plated at 104 cells/well at day 0 and the number of cells/well was monitored at daily intervals. At each time point, 3 wells of cells were counted and the number of cells is shown as average of the three samples. The culture media was changed at day 4. The _Brd2_−/− cells grew very poorly as compared to the wild-type cells, and the heterozygous Brd2+/− MEFs grew at an intermediate rate as compared to the Brd2+/+ and _Brd2_−/−cells. Panel B: Flow cytometry analysis of cell cycle of MEFs of E9.5 _Brd2_−/− (157-5) and Brd2+/+ (157-11) embryos, showing an elevated G1 peak (2N) in the mutant cells, indicating that the cell cycle of _Brd2_−/− MEFs is delayed at the G1 phase.
Figure 4
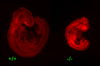
Lysotracker staining to detect apoptotic cells in E9.5 embryos. Embryos were dissected and processed for Lysotracker staining in the whole embryos (see materials and Methods). Photomicrographs of the entire embryos of Brd2+/+ versus _Brd2_−/− are shown. There are enhanced levels of positive lysotracker staining in the _Brd2_-deficient embryo.
Similar articles
- The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis.
Gyuris A, Donovan DJ, Seymour KA, Lovasco LA, Smilowitz NR, Halperin AL, Klysik JE, Freiman RN. Gyuris A, et al. Biochim Biophys Acta. 2009 May;1789(5):413-21. doi: 10.1016/j.bbagrm.2009.03.005. Epub 2009 Apr 10. Biochim Biophys Acta. 2009. PMID: 19362612 Free PMC article. - BET proteins are essential for the specification and maintenance of the epiblast lineage in mouse preimplantation embryos.
Tsume-Kajioka M, Kimura-Yoshida C, Mochida K, Ueda Y, Matsuo I. Tsume-Kajioka M, et al. BMC Biol. 2022 Mar 9;20(1):64. doi: 10.1186/s12915-022-01251-0. BMC Biol. 2022. PMID: 35264162 Free PMC article. - Change in nuclear-cytoplasmic localization of a double-bromodomain protein during proliferation and differentiation of mouse spinal cord and dorsal root ganglia.
Crowley T, Brunori M, Rhee K, Wang X, Wolgemuth DJ. Crowley T, et al. Brain Res Dev Brain Res. 2004 Apr 19;149(2):93-101. doi: 10.1016/j.devbrainres.2003.12.011. Brain Res Dev Brain Res. 2004. PMID: 15063089 - Bromodomain and Extraterminal Domain Protein 2 in Multiple Human Diseases.
Ji Y, Chen W, Wang X. Ji Y, et al. J Pharmacol Exp Ther. 2024 May 21;389(3):277-288. doi: 10.1124/jpet.123.002036. J Pharmacol Exp Ther. 2024. PMID: 38565308 Review. - The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins.
Taniguchi Y. Taniguchi Y. Int J Mol Sci. 2016 Nov 7;17(11):1849. doi: 10.3390/ijms17111849. Int J Mol Sci. 2016. PMID: 27827996 Free PMC article. Review.
Cited by
- Epigenetics.
Jain R, Epstein JA. Jain R, et al. Adv Exp Med Biol. 2024;1441:341-364. doi: 10.1007/978-3-031-44087-8_18. Adv Exp Med Biol. 2024. PMID: 38884720 Review. - BRD2 and BRD3 genes independently evolved RNA structures to control unproductive splicing.
Petrova M, Margasyuk S, Vorobeva M, Skvortsov D, Dontsova OA, Pervouchine DD. Petrova M, et al. NAR Genom Bioinform. 2024 Jan 15;6(1):lqad113. doi: 10.1093/nargab/lqad113. eCollection 2024 Mar. NAR Genom Bioinform. 2024. PMID: 38226395 Free PMC article. - Epigenetic regulation of craniofacial development and disease.
Shull LC, Artinger KB. Shull LC, et al. Birth Defects Res. 2024 Jan;116(1):e2271. doi: 10.1002/bdr2.2271. Epub 2023 Nov 14. Birth Defects Res. 2024. PMID: 37964651 Review. - Bromodomain (BrD) Family Members as Regulators of Cancer Stemness-A Comprehensive Review.
Czerwinska P, Mackiewicz AA. Czerwinska P, et al. Int J Mol Sci. 2023 Jan 4;24(2):995. doi: 10.3390/ijms24020995. Int J Mol Sci. 2023. PMID: 36674511 Free PMC article. Review. - Genome-Wide Identification and Characterization of the BRD Family in Nile Tilapia (Oreochromis niloticus).
Xu C, Yu M, Zhang Q, Ma Z, Du K, You H, Wei J, Wang D, Tao W. Xu C, et al. Animals (Basel). 2022 Sep 1;12(17):2266. doi: 10.3390/ani12172266. Animals (Basel). 2022. PMID: 36077987 Free PMC article.
References
- An FQ, Compitello N, Horwitz E, Sramkoski M, Knudsen ES, Renne R. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J Biol Chem. 2005;280:3862–3874. - PubMed
- Annegers JF. Epidemiology and genetics of epilepsy. Neurologic Clinics. 1994;12:15–29. - PubMed
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. - PubMed
- Beck S, Hanson I, Kelly A, Pappin DJ, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. - PubMed
- BelAiba RS, Baril P, Chebloune Y, Tabone E, Boukerche H. Identification and cloning of an 85-kDa protein homologous to RING3 that is upregulated in proliferating endothelial cells. Eur J Biochem. 2001;268:4398–4407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK031775-24/DK/NIDDK NIH HHS/United States
- GM 081767/GM/NIGMS NIH HHS/United States
- DK31775/DK/NIDDK NIH HHS/United States
- R01 NS027941-19S1/NS/NINDS NIH HHS/United States
- R01 NS027941/NS/NINDS NIH HHS/United States
- T32 DK07647/DK/NIDDK NIH HHS/United States
- NS27941/NS/NINDS NIH HHS/United States
- R01 DK031775/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous