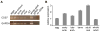A hypervariable invertebrate allodeterminant - PubMed (original) (raw)
A hypervariable invertebrate allodeterminant
Matthew L Nicotra et al. Curr Biol. 2009.
Abstract
Colonial marine invertebrates, such as sponges, corals, bryozoans, and ascidians, often live in densely populated communities where they encounter other members of their species as they grow over their substratum. Such encounters typically lead to a natural histocompatibility response in which colonies either fuse to become a single, chimeric colony or reject and aggressively compete for space. These allorecognition phenomena mediate intraspecific competition, support allotypic diversity, control the level at which selection acts, and resemble allogeneic interactions in pregnancy and transplantation. Despite the ubiquity of allorecognition in colonial phyla, however, its molecular basis has not been identified beyond what is currently known about histocompatibility in vertebrates and protochordates. We positionally cloned an allorecognition gene by using inbred strains of the cnidarian, Hydractinia symbiolongicarpus, which is a model system for the study of invertebrate allorecognition. The gene identified encodes a putative transmembrane receptor expressed in all tissues capable of allorecognition that is highly polymorphic and predicts allorecognition responses in laboratory and field-derived strains. This study reveals that a previously undescribed hypervariable molecule bearing three extracellular domains with greatest sequence similarity to the immunoglobulin superfamily is an allodeterminant in a lower metazoan.
Figures
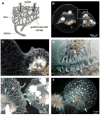
Figure 1
Allorecognition in Hydractinia symbiolongicarpus. (A) Morphology of a Hydractinia colony. (B) Fusion between two Hydractinia colonies. (C) Magnification of the boxed area in B. (D) Magnification of a stolon tip and stolon flank from rejecting colonies, showing recruitment of nematocytes (arrowheads) to the contact point. (E) Early rejection, showing production of hyperplastic stolons (arrowhead). (F) Late stage rejection, showing proliferation of hyperplastic stolons (arrowheads). D, E, and F show different colonies. A modified from [37]. B and C reprinted with permission from [21]. F from [26]. D from [14], reprinted with permission of John Wiley & Sons, Inc. © 1989.
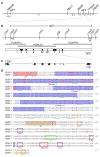
Figure 2
Positional cloning of alr2. (A) Genetic map of the Hydractinia allorecognition complex, which contains two histocompatibility loci, alr1 and alr2. 194m6, 18m1, 28m6, 174m4, and 29m9 are markers used to map the interval. Genetic distances are shown below the line in centimorgans. (B) Physical map of the alr2 genomic region. Markers are derived from fosmid or BAC sequences. Recombination breakpoints defining the alr2 locus are shown as X’s. BAC clones comprising the minimum tiling path across the alr2 region are shown as heavy gray lines. Numbered boxes are expressed coding sequences. (C) Genomic organization of CDS7. White boxes, black boxes, and bent lines indicate UTRs, exons, and introns, respectively. (D) CDS7 amino acid sequence, showing alignment between the f and r alleles. SP = signal peptide, domains I-III = regions similar to Ig-like domains, TM = transmembrane domain; PKC = protein kinase C phosphorylation motif, TKP = tyrosine kinase phosphorylation motif, CK2 = casein kinase II phosphorylation motif, EM = endocytosis motif, ITIM-like = ITIM-like motif. Potential N-glycosylation sites are underlined. Exon organization is denoted below the alignment. < and > = amino acids encoded by the last complete codon of the left and right exon ends, respectively. ^ = codon spanning two exons.
Figure 3
Expression of CDS7. (A) RT-PCR of cDNA isolated from different Hydractinia life stages and tissues. Top panel shows a 509 bp product amplified from exons 8–9 of CDS7. Bottom panel shows a 406 bp product amplified from GAPDH. −RT = control in which reverse transcriptase was omitted from cDNA synthesis. (B) Real-time PCR of CDS7. Relative expression shown for eggs, 2–8 cell embryos (early blastula), 64 cell embryos (late blastula), planulae, adult mat tissue, and whole adult colonies (polyps + mat).
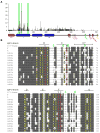
Figure 4
Variability of CDS7. (A) High levels of per-site variability suggest positive selection for diversity over CDS7. Variability was calculated as (number of different residues)/(frequency of the most common residue) − 1. Triangles and green bars indicate sites demonstrating significant departures from neutrality, suggesting positive selection. CDS7 domains labeled as in Figure 2. (B) PRANKF [25] alignment of domain I from 16 CDS7 alleles. Eight or more consensus residues are shaded gray. Green arrowheads indicate positions under positive selection. Residues matching the conserved V/I-frame are in yellow, those that differ are in red with V/I-frame consensus residues shown above. Beta-strand positions in canonical Ig-like domains relative to the putative V/I-frame residues are shown above the alignment.
Comment in
- Invertebrate allorecognition: the origins of histocompatibility.
Dishaw LJ, Litman GW. Dishaw LJ, et al. Curr Biol. 2009 Apr 14;19(7):R286-8. doi: 10.1016/j.cub.2009.02.035. Curr Biol. 2009. PMID: 19368870 Free PMC article.
Similar articles
- A family of unusual immunoglobulin superfamily genes in an invertebrate histocompatibility complex.
Huene AL, Sanders SM, Ma Z, Nguyen AD, Koren S, Michaca MH, Mullikin JC, Phillippy AM, Schnitzler CE, Baxevanis AD, Nicotra ML. Huene AL, et al. Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2207374119. doi: 10.1073/pnas.2207374119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161920 Free PMC article. - Invertebrate allorecognition: the origins of histocompatibility.
Dishaw LJ, Litman GW. Dishaw LJ, et al. Curr Biol. 2009 Apr 14;19(7):R286-8. doi: 10.1016/j.cub.2009.02.035. Curr Biol. 2009. PMID: 19368870 Free PMC article. - Allorecognition proteins in an invertebrate exhibit homophilic interactions.
Karadge UB, Gosto M, Nicotra ML. Karadge UB, et al. Curr Biol. 2015 Nov 2;25(21):2845-2850. doi: 10.1016/j.cub.2015.09.030. Epub 2015 Oct 8. Curr Biol. 2015. PMID: 26455308 Free PMC article. - Model systems of invertebrate allorecognition.
Rosengarten RD, Nicotra ML. Rosengarten RD, et al. Curr Biol. 2011 Jan 25;21(2):R82-92. doi: 10.1016/j.cub.2010.11.061. Curr Biol. 2011. PMID: 21256442 Review. - The Hydractinia allorecognition system.
Nicotra ML. Nicotra ML. Immunogenetics. 2022 Feb;74(1):27-34. doi: 10.1007/s00251-021-01233-6. Epub 2021 Nov 13. Immunogenetics. 2022. PMID: 34773127 Review.
Cited by
- A family of unusual immunoglobulin superfamily genes in an invertebrate histocompatibility complex.
Huene AL, Sanders SM, Ma Z, Nguyen AD, Koren S, Michaca MH, Mullikin JC, Phillippy AM, Schnitzler CE, Baxevanis AD, Nicotra ML. Huene AL, et al. Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2207374119. doi: 10.1073/pnas.2207374119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161920 Free PMC article. - Genetic Background and Allorecognition Phenotype in Hydractinia symbiolongicarpus.
Powell AE, Moreno M, Gloria-Soria A, Lakkis FG, Dellaporta SL, Buss LW. Powell AE, et al. G3 (Bethesda). 2011 Nov;1(6):499-504. doi: 10.1534/g3.111.001149. Epub 2011 Nov 1. G3 (Bethesda). 2011. PMID: 22384360 Free PMC article. - High amino acid diversity and positive selection at a putative coral immunity gene (tachylectin-2).
Hayes ML, Eytan RI, Hellberg ME. Hayes ML, et al. BMC Evol Biol. 2010 May 19;10:150. doi: 10.1186/1471-2148-10-150. BMC Evol Biol. 2010. PMID: 20482872 Free PMC article. - The colonial cnidarian Hydractinia.
Frank U, Nicotra ML, Schnitzler CE. Frank U, et al. Evodevo. 2020 Mar 26;11:7. doi: 10.1186/s13227-020-00151-0. eCollection 2020. Evodevo. 2020. PMID: 32226598 Free PMC article. Review. - Stem Cells and Innate Immunity in Aquatic Invertebrates: Bridging Two Seemingly Disparate Disciplines for New Discoveries in Biology.
Ballarin L, Karahan A, Salvetti A, Rossi L, Manni L, Rinkevich B, Rosner A, Voskoboynik A, Rosental B, Canesi L, Anselmi C, Pinsino A, Tohumcu BE, Jemec Kokalj A, Dolar A, Novak S, Sugni M, Corsi I, Drobne D. Ballarin L, et al. Front Immunol. 2021 Jun 30;12:688106. doi: 10.3389/fimmu.2021.688106. eCollection 2021. Front Immunol. 2021. PMID: 34276677 Free PMC article. Review.
References
- Francis L. Intraspecific aggression and its effect on the distribution of Anthopleura elegantissima and some related sea anemones. Biol Bull. 1973;144:73–92. - PubMed
- Buss LW, Grosberg RK. Morphogenetic basis for phenotypic differences in hydroid competitive behaviour. Nature. 1990;343:63–66.
- Buss LW. Competition within and between encrusting clonal invertebrates. Trends Ecol Evol. 1990;5:352–356. - PubMed
- Grosberg RK. The evolution of allorecognition specificity in clonal invertebrates. Q Rev Biol. 1988;63:377–412.
Publication types
MeSH terms
Grants and funding
- R21 AI066242/AI/NIAID NIH HHS/United States
- T32 GM007499/GM/NIGMS NIH HHS/United States
- T32-GM07499/GM/NIGMS NIH HHS/United States
- 1R21-AI066242/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources