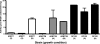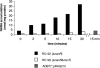Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase - PubMed (original) (raw)
Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase
Christopher Brigham et al. J Bacteriol. 2009 Jun.
Abstract
We characterized the nanLET operon in Bacteroides fragilis, whose products are required for the utilization of the sialic acid N-acetyl neuraminic acid (NANA) as a carbon and energy source. The first gene of the operon is nanL, which codes for an aldolase that cleaves NANA into N-acetyl mannosamine (manNAc) and pyruvate. The next gene, nanE, codes for a manNAc/N-acetylglucosamine (NAG) epimerase, which, intriguingly, possesses more similarity to eukaryotic renin binding proteins than to other bacterial NanE epimerase proteins. Unphosphorylated manNAc is the substrate of NanE, while ATP is a cofactor in the epimerase reaction. The third gene of the operon is nanT, which shows similarity to the major transporter facilitator superfamily and is most likely to be a NANA transporter. Deletion of any of these genes eliminates the ability of B. fragilis to grow on NANA. Although B. fragilis does not normally grow with manNAc as the sole carbon source, we isolated a B. fragilis mutant strain that can grow on this substrate, likely due to a mutation in a NAG transporter; both manNAc transport and NAG transport are affected in this strain. Deletion of the nanE epimerase gene or the rokA hexokinase gene, whose product phosphorylates NAG, in the manNAc-enabled strain abolishes growth on manNAc. Thus, B. fragilis possesses a new pathway of NANA utilization, which we show is also found in other Bacteroides species.
Figures
FIG. 1.
Schematic of the nanLET/nanR operon. The nanR gene product is a ROK family repressor protein. The nanL gene product is a NANA lyase (aldolase). The nanE gene product possesses similarity to the mammalian RnBP, also known to be an _N_-acetylglucosamine 2-epimerase. The nanT gene product is a transport protein of the major facilitator superfamily. The hyp gene encodes a hypothetical protein. Relevant deletion constructs and the schematics of the deletions are listed.
FIG. 2.
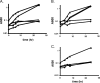
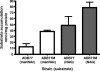
Growth phenotypes of nanR and nanLET gene knockout strains, and manNAc-utilizing strains. (A) Strains were grown in SAMM broth containing thymine, 0.5% NANA, and 0.02% glucose as described in Materials and Methods. Strains: ADB77 (wild type, ▪) and RC122 (Δ_nanR_, ▴), ΔL1 (Δ_nanL_, ▾), RC201 (Δ_nanE_, ⧫), RC140 (Δ_nanT_, •). (B) Growth on manNAc. Strains were inoculated in SAMM with thymine and 0.5% manNAc as described in Materials and Methods. Strains: ADB77 (▪), ADB77M (▴), ADB77MΔL1 (▾), ADB77MΔT (•), ADB77MΔE (⧫). (C) Growth of manNAc-utilizing strain and derivatives on NANA. Symbols represent the same strains as in panel B. Strains were inoculated into SAMM with thymine, 0.5% NANA, and 0.02% glucose as described in Materials and Methods. All curves shown here are representative of three separate growth experiments.
FIG. 3.
NANA lyase (aldolase) activities of B. fragilis strains. All strains were grown in SAMM broth with thymine and 0.5% of the indicated sugar. Extracts were prepared from cells and NANA lyase (aldolase) activity was measured as indicated in the supplemental material. ADB77 (wild type) cells were grown in xylose (X), glucose (G), and NANA (N). ADB77M cells were grown in glucose, NANA, and manNAc (M). RC122 (Δ_nanR_) cells were grown in xylose, glucose, and NANA.
FIG. 4.
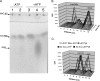
NanE enzymatic activity. (A) manNAc is converted to NAG in an ATP-dependent manner. Extracts of B. fragilis cells with (ADB77, lanes 2 and 4) or without (RC201, lanes 1 and 3) an intact nanE gene were incubated with manNAc with or without ATP as described in Materials and Methods. Lane 5 is manNAc incubated in buffer without added extract. Arrows indicate the positions of NAG, manNAc, and NAG6-P standards, as determined in separate experiments. (B) manNAc→NAG steady-state ratio is 1:3. Purified NanEHis6 was incubated with ATP and [14C]manNAc (specific activity, 20 μCi/mmol) for the times indicated. (C) ATP, but not ATP, hydrolysis is required for manNAc epimerase activity. Purified NanEHis6 was incubated with or without ATP or ATP-γ-P and [14C]manNAc (specific activity, 20 μCi/mmol) as indicated. Each reaction in panel C was incubated for 18 h at 37°C. In panels B and C, the positions of manNac and NAG were determined in separate experiments.
FIG. 5.
NANA accumulation in RC122, RC122T, and the nanL disruption strain ADB77::pRAG210. Cells were incubated with labeled sugar (specific activity, 10 μCi/mmol) for a time course of 30 min as described in Materials and Methods. Aliquots were harvested at the times indicated on the x axis. The [14C]NANA specific activity was 0.1 μCi/mmol. The experiment with ADB77::pRAG210 cells was incubated for 15 min as described in Materials and Methods.
FIG. 6.
manNAc and NAG uptake in ADB77 (wild type) and ADB77M. Cells were incubated with labeled sugar (specific activity, 2.5 μCi/mmol) as described in Materials and Methods. Accumulation experiments were performed in triplicate.
FIG. 7.
Comparison of the NANA and amino sugar utilization pathways of E. coli (A) and B. fragilis (B). (A) Pathway adapted from Vimr et al. (36). GlcN,
d
-glucosamine. (B) Enzymes in boldface have been assayed by our laboratory. The NagA reaction has not been experimentally verified by us; however, there is an annotated sequence of such a gene in the B. fragilis 638R partial genome sequence. X designates the transporter, probably a NAG transporter, that contains the enabling mutation present in the strain ADB77M.
FIG. 8.
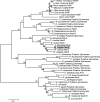
Phylogenetic tree of NanE proteins and RnBPs, including B. fragilis NanE (boxed). The tree was constructed by using the neighbor-joining method and was visualized with MEGA 3.1. hyp prot, hypothetical protein.
Similar articles
- Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli.
Plumbridge J, Vimr E. Plumbridge J, et al. J Bacteriol. 1999 Jan;181(1):47-54. doi: 10.1128/JB.181.1.47-54.1999. J Bacteriol. 1999. PMID: 9864311 Free PMC article. - Production of N-Acetyl-d-neuraminic Acid by Whole Cells Expressing Bacteroides thetaiotaomicron N-Acetyl-d-glucosamine 2-Epimerase and Escherichia coli N-Acetyl-d-neuraminic Acid Aldolase.
Gao X, Zhang F, Wu M, Wu Z, Shang G. Gao X, et al. J Agric Food Chem. 2019 Jun 5;67(22):6285-6291. doi: 10.1021/acs.jafc.9b01839. Epub 2019 May 28. J Agric Food Chem. 2019. PMID: 31117501 - The first committed step in the biosynthesis of sialic acid by Escherichia coli K1 does not involve a phosphorylated N-acetylmannosamine intermediate.
Ringenberg MA, Steenbergen SM, Vimr ER. Ringenberg MA, et al. Mol Microbiol. 2003 Nov;50(3):961-75. doi: 10.1046/j.1365-2958.2003.03741.x. Mol Microbiol. 2003. PMID: 14617154 - Regulation and pathophysiological implications of UDP-GlcNAc 2-epimerase/ManNAc kinase (GNE) as the key enzyme of sialic acid biosynthesis.
Reinke SO, Lehmer G, Hinderlich S, Reutter W. Reinke SO, et al. Biol Chem. 2009 Jul;390(7):591-9. doi: 10.1515/BC.2009.073. Biol Chem. 2009. PMID: 19426133 Review. - Functions, structures, and applications of cellobiose 2-epimerase and glycoside hydrolase family 130 mannoside phosphorylases.
Saburi W. Saburi W. Biosci Biotechnol Biochem. 2016 Jul;80(7):1294-305. doi: 10.1080/09168451.2016.1166934. Epub 2016 Mar 31. Biosci Biotechnol Biochem. 2016. PMID: 27031293 Review.
Cited by
- Host-Derived Sialic Acids Are an Important Nutrient Source Required for Optimal Bacterial Fitness In Vivo.
McDonald ND, Lubin JB, Chowdhury N, Boyd EF. McDonald ND, et al. mBio. 2016 Apr 12;7(2):e02237-15. doi: 10.1128/mBio.02237-15. mBio. 2016. PMID: 27073099 Free PMC article. - Comparison of gnotobiotic communities reveals milk-adapted metabolic functions and unexpected amino acid metabolism by the pre-weaning microbiome.
Lubin JB, Silverman MA, Planet PJ. Lubin JB, et al. Gut Microbes. 2024 Jan-Dec;16(1):2387875. doi: 10.1080/19490976.2024.2387875. Epub 2024 Aug 12. Gut Microbes. 2024. PMID: 39133869 Free PMC article. - Enhancement of Edwardsiella piscicida infection, biofilm formation, and motility caused by N-acetylneuraminate lyase.
Vo LK, Tran NT, Kubo Y, Sahashi D, Komatsu M, Shiozaki K. Vo LK, et al. Glycoconj J. 2022 Jun;39(3):429-442. doi: 10.1007/s10719-022-10045-z. Epub 2022 Feb 22. Glycoconj J. 2022. PMID: 35192095 - Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by porphyromonas gingivalis.
Aruni AW, Roy F, Fletcher HM. Aruni AW, et al. Infect Immun. 2011 Oct;79(10):3872-86. doi: 10.1128/IAI.05631-11. Epub 2011 Aug 8. Infect Immun. 2011. PMID: 21825062 Free PMC article. - A comparative study of the substrate preference of the sialidases, CpNanI, HpNanH, and BbSia2 towards 2-Aminobenzamide-labeled 3'-Sialyllactose, 6'-Sialyllactose, and Sialyllacto-N-tetraose-b.
Lata M, Ramya TNC. Lata M, et al. Biochem Biophys Rep. 2024 Jul 19;39:101791. doi: 10.1016/j.bbrep.2024.101791. eCollection 2024 Sep. Biochem Biophys Rep. 2024. PMID: 39156723 Free PMC article.
References
- Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102439-469. - PubMed
- Bravo, I. G., S. Garcia-Vallve, A. Romeu, and A. Reglero. 2004. Prokaryotic origin of cytidylyltransferases and alpha-ketoacid synthases. Trends Microbiol. 12120-128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases