Global mapping of protein-DNA interactions in vivo by digital genomic footprinting - PubMed (original) (raw)
doi: 10.1038/nmeth.1313. Epub 2009 Mar 22.
Xiaoyu Chen, Zhihong Zhang, Peter J Sabo, Richard Sandstrom, Alex P Reynolds, Robert E Thurman, Shane Neph, Michael S Kuehn, William S Noble, Stanley Fields, John A Stamatoyannopoulos
Affiliations
- PMID: 19305407
- PMCID: PMC2668528
- DOI: 10.1038/nmeth.1313
Global mapping of protein-DNA interactions in vivo by digital genomic footprinting
Jay R Hesselberth et al. Nat Methods. 2009 Apr.
Abstract
The orchestrated binding of transcriptional activators and repressors to specific DNA sequences in the context of chromatin defines the regulatory program of eukaryotic genomes. We developed a digital approach to assay regulatory protein occupancy on genomic DNA in vivo by dense mapping of individual DNase I cleavages from intact nuclei using massively parallel DNA sequencing. Analysis of >23 million cleavages across the Saccharomyces cerevisiae genome revealed thousands of protected regulatory protein footprints, enabling de novo derivation of factor binding motifs and the identification of hundreds of new binding sites for major regulators. We observed striking correspondence between single-nucleotide resolution DNase I cleavage patterns and protein-DNA interactions determined by crystallography. The data also yielded a detailed view of larger chromatin features including positioned nucleosomes flanking factor binding regions. Digital genomic footprinting should be a powerful approach to delineate the cis-regulatory framework of any organism with an available genome sequence.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
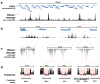
Fig. 1. Digital DNase I analysis of yeast chromatin structure from chromosomal to nucleotide resolution
(a) Per-nucleotide DNase I cleavage density across an exemplary 100-kilobase region of chromosome 10 (chr10:625,000-725,000) containing ∼50 open reading frames (ORFs). (b) Magnification of exemplary ∼2.5 kb regions containing RSM17/YJR115W and ECM17/IML1 intergenic intervals. (c) Further magnification showing positions of individual DNase I cleavage events (stacked vertical black tick marks), revealing short regions protected from DNase I cleavage (DNase I “footprints”). (d) Resolution of individual DNase I footprints (red shading) with known motifs for yeast regulatory factors Rap1, Reb1, Abf1 and Cbf1. The dashed black line indicates the average level of DNase I cleavage throughout the genome (avg. ∼2 cleavages per bp).
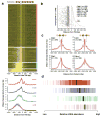
Fig. 2. Detection of footprints and corresponding sequence motifs
(a) Visualization of DNase I protection (footprinting) around 907 computationally-predicted Reb1 sites in a heat map. Rows show levels of DNase I cleavage 25 bp up- and downstream of each motif instance and are sorted by the ratio of mean cleavage over flanking regions to that within the motif itself. Red ticks (at left) indicate motif instances (n = 580) that coincide with footprints (FDR = 0.05) containing _de novo_-derived Reb1 motifs. Blue ticks (right) indicate motif instances (n = 151) coinciding with those identified by ChIP. All motif instances are uniquely mappable within the yeast genome. (b) Mean per nucleotide DNase I cleavage (red) and evolutionary conservation (Phastcons?; blue) calculated for footprints that match the Reb1, Abf1, Rap1 and Hsf1 motifs (subpanel vertical axes). Significance of observed conservation patterns (blue text) (Supplementary Methods), extent of consensus motifs derived from the footprinted region (green shading), motifs derived from ChIP and footprinting below. Venn diagrams depict the overlap of motifs derived from and mapping to footprints (red) vs. ChIP (blue).
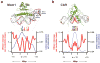
Fig. 3. Mean nucleotide-level accessibility parallels protein:DNA interactions
(a) Structure of Mcm1 (green) bound to a single DNA recognition site (adjacent Matα2 was removed for clarity). Colored DNA bases correspond to positions within the footprint-derived Mcm1 motif (below), and red DNA backbone coloration reflects the extent of observed DNase I cleavage across 88 Mcm1 sites (red trace in subpanel). Mean nucleotide-level conservation by PhastCons is shown in parallel (blue trace in subpanel; P < 10-5). (b) Structure of the human homolog of CBF1 is shown relative to the mean nucleotide level cleavage and conservation (P < 10-3) across 243 Cbf1 sites.
Fig. 4. Individual yeast regulatory regions and factor binding sites
(a) Rap1 binds to two adjacent sites also predicted from ChIP experiments upstream of RPS6A (chr16:378,775-378,874). (b) Reb1 binds to a canonical site upstream of TUF1 (chr15:683,707-683,806) but a non-canonical site upstream is only inferred from ChIP data (c) Mcm1 site upstream of MFA1 (chr4:1,384,893-1,384,993) exhibits hypersensitive nucleotides illustrated in Fig. 3a. (d) Hsf1 site identified by ChIP in BTN2 promoter (chr7:772,068-772,167) is identified as a footprint. (e) Two Reb1 binding sites in the REB1 promoter (chr2:336,885-337,084) are identified as footprints; a Cbf1 site predicted by ChIP shows a footprint, but a Rpn4 site defined by ChIP does not. (f) Two Pdr3 sites in the PDR5 promoter (chr15:619,227-619,476) are identified as footprints, in addition to an evolutionarily conserved region further upstream. Each panel shows per nucleotide DNase I cleavage, detected footprints (red boxes), assigned motifs (pink boxes), binding sites inferred from ChIP experiments (blue boxes), and evolutionary conservation (dark blue, Phastcons, bottom).
Fig. 5. Higher-order patterns of DNA accessibility
(a) Mapped DNase I cleavages relative to 5,006 TSSs. Four major clusters are exposed by _k_-means analysis (red, blue, green and purple bars, respectively). In the red cluster, maximal DNase I cleavage occurs in a stereotypic ∼50 bp band ∼100 bp upstream of the TSS (grey arrowhead, top). In the blue, green and purple clusters, the extent and intensity of DNase I cleavage upstream of the TSS widens to the -1, -2, and -3 nucleosomes (respectively). (b) Spatial restriction of footprints near TSSs. Distribution of footprints matching Reb1, Abf1, Rap1, Mcm1, Pdr3, Cbf1 and Hsf1 relative to the TSSs (dashed black lines) and start codons of 1,260 genes sorted by the length of the 5′UTR. Enrichment within a ∼50 bp region centered ∼100 bp upstream of the TSS (dashed red lines). (c) DNase I cleavage profiles aligned relative to Reb1, Abf1, Rap1 and Mcm1 footprints. (d) mRNA abundance for genes found in each of the four clusters correlates with the accessibility of the promoters of those genes (colors as in a; median expression denoted by a black bar).
Similar articles
- Footprints by deep sequencing.
Hager G. Hager G. Nat Methods. 2009 Apr;6(4):254-5. doi: 10.1038/nmeth0409-254. Nat Methods. 2009. PMID: 19333240 No abstract available. - High-resolution mapping of in vivo genomic transcription factor binding sites using in situ DNase I footprinting and ChIP-seq.
Chumsakul O, Nakamura K, Kurata T, Sakamoto T, Hobman JL, Ogasawara N, Oshima T, Ishikawa S. Chumsakul O, et al. DNA Res. 2013 Aug;20(4):325-38. doi: 10.1093/dnares/dst013. Epub 2013 Apr 11. DNA Res. 2013. PMID: 23580539 Free PMC article. - Genomic footprinting.
Vierstra J, Stamatoyannopoulos JA. Vierstra J, et al. Nat Methods. 2016 Mar;13(3):213-21. doi: 10.1038/nmeth.3768. Nat Methods. 2016. PMID: 26914205 Review. - Survey of protein-DNA interactions in Aspergillus oryzae on a genomic scale.
Wang C, Lv Y, Wang B, Yin C, Lin Y, Pan L. Wang C, et al. Nucleic Acids Res. 2015 May 19;43(9):4429-46. doi: 10.1093/nar/gkv334. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883143 Free PMC article. - A survey of DNA motif finding algorithms.
Das MK, Dai HK. Das MK, et al. BMC Bioinformatics. 2007 Nov 1;8 Suppl 7(Suppl 7):S21. doi: 10.1186/1471-2105-8-S7-S21. BMC Bioinformatics. 2007. PMID: 18047721 Free PMC article. Review.
Cited by
- Lagging-strand replication shapes the mutational landscape of the genome.
Reijns MAM, Kemp H, Ding J, de Procé SM, Jackson AP, Taylor MS. Reijns MAM, et al. Nature. 2015 Feb 26;518(7540):502-506. doi: 10.1038/nature14183. Epub 2015 Jan 26. Nature. 2015. PMID: 25624100 Free PMC article. - The chromatin fingerprint of gene enhancer elements.
Zentner GE, Scacheri PC. Zentner GE, et al. J Biol Chem. 2012 Sep 7;287(37):30888-96. doi: 10.1074/jbc.R111.296491. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952241 Free PMC article. Review. - Surveying the epigenomic landscape, one base at a time.
Zentner GE, Henikoff S. Zentner GE, et al. Genome Biol. 2012 Oct 22;13(10):250. doi: 10.1186/gb4051. Genome Biol. 2012. PMID: 23088423 Free PMC article. Review. - Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models.
Chen J, Lalonde S, Obrdlik P, Noorani Vatani A, Parsa SA, Vilarino C, Revuelta JL, Frommer WB, Rhee SY. Chen J, et al. Front Plant Sci. 2012 Jun 21;3:124. doi: 10.3389/fpls.2012.00124. eCollection 2012. Front Plant Sci. 2012. PMID: 22737156 Free PMC article. - Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis.
Zhang W, Zhang T, Wu Y, Jiang J. Zhang W, et al. Plant Cell. 2012 Jul;24(7):2719-31. doi: 10.1105/tpc.112.098061. Epub 2012 Jul 5. Plant Cell. 2012. PMID: 22773751 Free PMC article.
References
- Maniatis T, Ptashne M. Structure of the lambda operators. Nature. 1973;246:133–6. - PubMed
- Gilbert W. Polymerization in Biological Systems. Elsevier; North-Holland, Amsterdam: 1972. pp. 245–259.
- Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. - PubMed
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM071923/GM/NIGMS NIH HHS/United States
- U54HG004592/HG/NHGRI NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- R01GM071923/GM/NIGMS NIH HHS/United States
- P41 RR011823-135975/RR/NCRR NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U54 HG004592/HG/NHGRI NIH HHS/United States
- P41RR11823/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases