Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection - PubMed (original) (raw)
Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection
Sebla B Kutluay et al. J Virol. 2009 Jun.
Abstract
During lytic infection by herpes simplex virus type 1 (HSV-1), histones are present at relatively low levels on the viral genome. However, the mechanisms that account for such low levels--how histone deposition on the viral genome is blocked or how histones are removed from the genome--are not yet defined. In this study, we show that histone occupancy on the viral genome gradually increased with time when transcription of the viral immediate-early (IE) genes was inhibited either by deletion of the VP16 activation domain or by chemical inhibition of RNA polymerase II (RNAP II). Inhibition of IE protein synthesis by cycloheximide did not affect histone occupancy on most IE promoters and coding regions but did cause an increase at delayed-early and late gene promoters. IE gene transcription from HSV-1 genomes associated with high levels of histones was stimulated by superinfection with HSV-2 without altering histone occupancy or covalent histone modifications at IE gene promoters. Moreover, RNAP II and histones cooccupied the viral genome in this context, indicating that RNAP II does not preferentially associate with viral genomes that are devoid of histones. These results suggest that during lytic infection, VP16, RNAP II, and IE proteins may all contribute to the low levels of histones on the viral genome, and yet the dearth of histones is neither a prerequisite for nor a necessary result of VP16-dependent transcription of nucleosomal viral genomes.
Figures
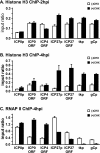
FIG. 1.
Core histone occupancy on the viral genome increases in RP5 lytic infections. HeLa cells were infected with the KOS (K) or RP5 (R) strain of HSV-1 at an MOI of 1 PFU/cell or 0.005 PFU/cell, respectively. ChIP was performed to detect the occupancy of histones H2A (A), H2B (B), H3 (C), and H4 (D) and RNAP II (E) on ICP27, ICP0, tk, and gC promoters (pro) and on the ICP27 ORF at 2, 4, and 6 hpi. The data were analyzed as explained in Materials and Methods. The graphs show the results of a representative experiment; similar results were observed for replicate experiments. IFN-Bp, IFN-β promoter; U3p, U3 snRNA promoter.
FIG. 2.
Inhibition of transcription by actinomycin D increases the core histone occupancy on the HSV-1 genome. HeLa cells were infected with HSV-1 strain KOS at an MOI of 1 PFU/cell in the presence (+) or absence (−) of actinomycin D (1 μg/ml). ChIP assays were performed to detect the occupancy of histone H2A (A), H2B (B), H3 (C), RNA polymerase II (D), and VP16 (E) on the ICP27, ICP1, tk, and gC promoters (pro) and on the ICP27 ORF at 2, 4, and 6 hpi. (A to C) Input ratios were calculated as follows. The percent input [% input (IP-noab)] values obtained for viral DNA fragments (after subtracting the values for the control samples without primary antibody) were normalized against the values for a cellular control gene fragment (IFN-β promoter [IFN-βp]). (D) Percent input values represent signals obtained from immunoprecipitated samples after subtracting background signals obtained in the absence of antibodies, expressed as a fraction of the signals obtained using input DNA. Graphs show the results of a representative experiment; similar results were observed in independent replicates.
FIG. 3.
Changes in histone occupancy on the HSV-1 genome upon inhibition of IE protein synthesis by cycloheximide. HeLa cells were pretreated with 60 μg/ml cycloheximide for 2 h and then were infected with HSV-1 KOS at an MOI of 1 PFU/cell in the presence of cycloheximide [(+)CHX]. (A) At 2 hpi, chromatin was cross-linked, and a ChIP assay probing histone H3 on the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), the ICP0 ORF, the ICP4 ORF, the ICP27 ORF, the tk promoter (tkp), and the gC promoter (gCp) was performed as indicated before. (B and C) At 4 hpi, chromatin was cross-linked, and a ChIP assay probing histone H3 (B) and RNAP II (C) on the indicated viral gene fragments was performed as indicated in the legend to Fig. 2. The data shown represent the averages from two independent experiments; the error bars indicate the ranges between the averages from these experiments.
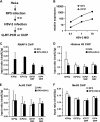
FIG. 4.
HSV-2 superinfection does not cause major changes in histone occupancy or covalent histone modifications on the RP5 genome. (A) Summary of the assays performed. (B) HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 0.1, 1, or 10 PFU/cell. At 2 h after HSV-2 superinfection, levels of ICP4 and ICP27 expression were analyzed by qRT-PCR. (C to F) HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 10 PFU/cell. At 2 h after HSV-2 superinfection, ChIP was performed, assaying the presence of RNAP II (C), histone H3 (D), H3K9/K14ac (E), and H3K4me3 (F) on the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), the ICP4 ORF, and the ICP27 ORF. The data shown represent the averages from three independent experiments, where percent input values from viral genes are normalized against those from the U3 snRNA promoter (U3p). Error bars represent the standard deviations for the averages from these experiments.
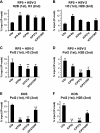
FIG. 5.
Histone and RNAP II cooccupancy on RP5 IE genes upon HSV-2 superinfection. HeLa cells were infected with RP5 at an MOI of 0.001 PFU/cell and, at 6 hpi, were superinfected with HSV-2 at an MOI of 10 PFU/cell. At 2 h after HSV-2 superinfection, seq-ChIP was performed, assaying the cooccupancy of histone H3, histone H2B, and RNAP II on the U3 snRNA promoter (U3p), the IFN-β promoter (IFN-Bp), the ICP0 promoter (ICP0p), the ICP27 promoter (ICP27p), and the ICP27 ORF. (A) Histone H2B (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (B) Histone H3 (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. (C) RNAP II (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (D) RNAP II (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. (E) HeLa cells were infected with KOS at an MOI of 1 PFU/cell, and seq-ChIP was performed at 2 hpi, as described above. RNAP II (first immunoprecipitation) and H3 (second immunoprecipitation) cooccupancy. (F) seq-ChIP was performed with HeLa cells infected as described for panel E. RNAP II (first immunoprecipitation) and H2B (second immunoprecipitation) cooccupancy. The data shown in panels A, C, and D represent the averages from three independent experiments, with error bars representing the standard deviations between the averages from these experiments. The data shown in panel B represent the averages from two independent experiments, with error bars representing the ranges between the averages from these experiments. The data shown in panels E and F represent three independent immunoprecipitations done in parallel, where the error bars represent the standard deviations. Samples with mean values that vary significantly from those for the U3 snRNA promoters are indicated by asterisks (**, P ≤ 0.01; *, 0.01 ≤ P ≤ 0.05; paired Student's t test). Percent input [% input (IP-noab)] values represent signals obtained from immunoprecipitated samples after subtracting background signals obtained in the absence of antibodies, expressed as a fraction of the signals obtained using input DNA.
Similar articles
- Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity.
Kutluay SB, Doroghazi J, Roemer ME, Triezenberg SJ. Kutluay SB, et al. Virology. 2008 Apr 10;373(2):239-47. doi: 10.1016/j.virol.2007.11.028. Epub 2008 Jan 14. Virology. 2008. PMID: 18191976 Free PMC article. - Role of chromatin during herpesvirus infections.
Kutluay SB, Triezenberg SJ. Kutluay SB, et al. Biochim Biophys Acta. 2009 Jun;1790(6):456-66. doi: 10.1016/j.bbagen.2009.03.019. Epub 2009 Mar 31. Biochim Biophys Acta. 2009. PMID: 19344747 Free PMC article. Review. - [Research Advances in VP16 of the Herpes Virus].
Zhang Y, Chen A, Wang M. Zhang Y, et al. Bing Du Xue Bao. 2016 Nov;32(6):817-24. Bing Du Xue Bao. 2016. PMID: 30004657 Review. Chinese.
Cited by
- Role of polycomb proteins in regulating HSV-1 latency.
Watson Z, Dhummakupt A, Messer H, Phelan D, Bloom D. Watson Z, et al. Viruses. 2013 Jul 15;5(7):1740-57. doi: 10.3390/v5071740. Viruses. 2013. PMID: 23860385 Free PMC article. Review. - The checkpoints of viral gene expression in productive and latent infection: the role of the HDAC/CoREST/LSD1/REST repressor complex.
Roizman B. Roizman B. J Virol. 2011 Aug;85(15):7474-82. doi: 10.1128/JVI.00180-11. Epub 2011 Mar 30. J Virol. 2011. PMID: 21450817 Free PMC article. Review. - Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein.
Zalckvar E, Paulus C, Tillo D, Asbach-Nitzsche A, Lubling Y, Winterling C, Strieder N, Mücke K, Goodrum F, Segal E, Nevels M. Zalckvar E, et al. Proc Natl Acad Sci U S A. 2013 Aug 6;110(32):13126-31. doi: 10.1073/pnas.1305548110. Epub 2013 Jul 22. Proc Natl Acad Sci U S A. 2013. PMID: 23878222 Free PMC article. - Herpes simplex virus VP16, but not ICP0, is required to reduce histone occupancy and enhance histone acetylation on viral genomes in U2OS osteosarcoma cells.
Hancock MH, Cliffe AR, Knipe DM, Smiley JR. Hancock MH, et al. J Virol. 2010 Feb;84(3):1366-75. doi: 10.1128/JVI.01727-09. Epub 2009 Nov 25. J Virol. 2010. PMID: 19939931 Free PMC article. - Models of Herpes Simplex Virus Latency.
Canova PN, Charron AJ, Leib DA. Canova PN, et al. Viruses. 2024 May 8;16(5):747. doi: 10.3390/v16050747. Viruses. 2024. PMID: 38793628 Free PMC article. Review.
References
- Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 802063-2068. - PMC - PubMed
- Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 3011090-1093. - PubMed
- Black, J. C., J. E. Choi, S. R. Lombardo, and M. Carey. 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23809-818. - PubMed
- Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. - PubMed
- Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 1801-19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources