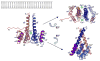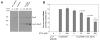The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae - PubMed (original) (raw)
The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae
Jan Abendroth et al. J Struct Biol. 2009 Jun.
Abstract
The type 2 secretion system (T2SS), a multi-protein machinery that spans both the inner and the outer membranes of Gram-negative bacteria, is used for the secretion of several critically important proteins across the outer membrane. Here we report the crystal structure of the N-terminal cytoplasmic domain of EpsF, an inner membrane spanning T2SS protein from Vibrio cholerae. This domain consists of a bundle of six anti-parallel helices and adopts a fold that has not been described before. The long C-terminal helix alpha6 protrudes from the body of the domain and most likely continues as the first transmembrane helix of EpsF. Two N-terminal EpsF domains form a tight dimer with a conserved interface, suggesting that the observed dimer occurs in the T2SS of many bacteria. Two calcium binding sites are present in the dimer interface with ligands provided for each site by both subunits. Based on this new structure, sequence comparisons of EpsF homologs and localization studies of GFP fused with EpsF, we propose that the second cytoplasmic domain of EpsF adopts a similar fold as the first cytoplasmic domain and that full-length EpsF, and its T2SS homologs, have a three-transmembrane helix topology.
Figures
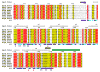
Figure 1. Family sequence alignment of the first cytoplasmic domain of EpsF and selected T2SS homologues
Identical residues are in red, similar residues according to the Risler matrix are in yellow. The predicted transmembrane helix (Met 171 – Val 192) is indicated with a green bar. The extent of the construct is marked with the black arrows above the sequences. The blue bar below the sequences indicates the solvent accessibility of each residue: dark blue is solvent exposed, white is buried. Residues involved in Ca2+-binding are marked with red triangles. Residues which are part of the dimer interface are marked with blue stars. Alpha-helical secondary structure symbols above the cyto1-EpsF sequence represent the observed structure in our experimental model.
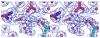
Figure 2. The dimer of cyto1-EpsF56-171 from V. cholerae
(A) Each cyto1-EpsF56-171 subunit consists of a six-helix bundle. The N-terminal residues have no secondary structure; the second half of the C-terminal helix protrudes away from the body of the domain and may extend towards the inner membrane as sketched in the left upper corner. Green spheres indicate calcium ions in the dimer interface. The dimer is shown in three different orientations related by 90° rotations about two different axes. (B) The interface of the cyto1-EpsF56-171 dimer. In this butterfly figure, the surfaces of the two cyto1-EpsF56-171 domains per dimer are colored in red and blue, respectively. Residues involved in dimer interactions are colored with the same color as the domain they contact. Light colors of the “imprint” indicate residues involved in hydrogen bonds, dark colors indicate residues involved in hydrogen bonds and hydrophobic interactions. Twelve hydrogen bonds in the dimer interface involve the following residues: (i) Glu 74 and Thr 77 provided by helix α1; (ii) Glu 95 from the loop between α2 and α3; and, (iii) Gln 161, Lys 162, Ser 165, Lys 166 and Glu 172, all belonging to the C-terminal part of helix α6. The following residues are involved in hydrophobic interactions: Leu 70, Arg 73, Gln 74, Thr 77, Leu 78, Gln 80 and Ser 81 provided by helix α1; Ala 91 from helix α2; and Lys 166, Gln 169, Ala 170, Leu 174 from helix α6. Clearly, most of the dimer interactions occur between equivalent helices α1 and α1′ and between α6 and α6′. (C) Conservation of interface residues according to the Consurf server (Landau et al., 2005)based on the sequence alignment of T2SS homologs of EpsF as shown in Figure 1. Comparison of Figures 2b and 2c shows that most dimer interactions involve residues from helices α1 and α6 and that interface residues are conserved. (Red: well conserved; blue less conserved).
Figure 2. The dimer of cyto1-EpsF56-171 from V. cholerae
(A) Each cyto1-EpsF56-171 subunit consists of a six-helix bundle. The N-terminal residues have no secondary structure; the second half of the C-terminal helix protrudes away from the body of the domain and may extend towards the inner membrane as sketched in the left upper corner. Green spheres indicate calcium ions in the dimer interface. The dimer is shown in three different orientations related by 90° rotations about two different axes. (B) The interface of the cyto1-EpsF56-171 dimer. In this butterfly figure, the surfaces of the two cyto1-EpsF56-171 domains per dimer are colored in red and blue, respectively. Residues involved in dimer interactions are colored with the same color as the domain they contact. Light colors of the “imprint” indicate residues involved in hydrogen bonds, dark colors indicate residues involved in hydrogen bonds and hydrophobic interactions. Twelve hydrogen bonds in the dimer interface involve the following residues: (i) Glu 74 and Thr 77 provided by helix α1; (ii) Glu 95 from the loop between α2 and α3; and, (iii) Gln 161, Lys 162, Ser 165, Lys 166 and Glu 172, all belonging to the C-terminal part of helix α6. The following residues are involved in hydrophobic interactions: Leu 70, Arg 73, Gln 74, Thr 77, Leu 78, Gln 80 and Ser 81 provided by helix α1; Ala 91 from helix α2; and Lys 166, Gln 169, Ala 170, Leu 174 from helix α6. Clearly, most of the dimer interactions occur between equivalent helices α1 and α1′ and between α6 and α6′. (C) Conservation of interface residues according to the Consurf server (Landau et al., 2005)based on the sequence alignment of T2SS homologs of EpsF as shown in Figure 1. Comparison of Figures 2b and 2c shows that most dimer interactions involve residues from helices α1 and α6 and that interface residues are conserved. (Red: well conserved; blue less conserved).
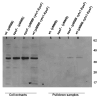
Figure 3. In vivo oligomerization of cyto1-EpsF
Wild type and epsF mutant strains of V. cholerae containing either pMMB67 or pMMB-cyto1-EpsF were grown in LB in the presence of IPTG to induce expression of cyto1-EpsF. Following growth, intact cells were incubated with increasing concentrations of DSP for 60 min. Then, the cells were centrifuged and subjected to SDS-PAGE and immunoblot analysis using anti-EpsF antibodies. The position of native EpsF and monomeric and cross-linked dimeric forms of cyto1-EpsF is indicated on the left. Molecular mass markers are shown on the right.
Figure 4. Cyto1-EpsF inhibits secretion of protease via the T2SS in V. cholerae
Wild type V. cholerae containing either pMMB67 as negative control or pMMB-cyto1-EpsF was grown in LB in the absence or presence of increasing concentrations of IPTG to induce the expression of cyto1-EpsF. Culture supernatants and cells were separated by centrifugation. (A) Cells were disrupted and subjected to SDS-PAGE and immunoblotting with anti-EpsF antibodies to determine the relative level of cyto1-EpsF expression at different IPTG concentrations. The position of molecular mass markers is shown on the left and native EpsF and cyto1-EpsF are indicated on the right. (B) Culture supernatants were tested for the presence of extracellular protease. The rate of hydrolysis was obtained from three independent experiments and the results are presented as percent extracellular activity ± standard error (SEM). The extracellular activity in the supernatant from the negative control was set to 100%. The epsF mutant, which is unable to secrete the protease, is shown as positive control.
Figure 5. Interaction of cyto1-EpsF and EpsL
EpsL was co-purified with cyto1-EpsFhis6 from V. cholerae cell extracts by metal-affinity chromatography and subjected to SDS-PAGE and immunoblotting with anti-EpsL antibodies. Lanes 1-4 show the level of EpsL detected in cell extracts prior to purification, and lanes 5-8 represent EpsL proteins that bound to hexahistidine-tagged cyto1-EpsF. No EpsL was purified when cyto1-EpsF was not expressed (lanes 5 and 6). Positions of molecular weight markers are shown.
Figure 6. Calcium binding site in the dimer interface of V. cholerae cyto1-EpsF56-171
Calcium is coordinated as an approximate tetragonal bipyramid by Glu 97 from chain A, and Glu 151 and Asp 155 from chain B, and three water molecules. Asp 155 is binding the calcium ion in a bidentate manner. Arg 73 forms a salt bridge with both Glu 151 and Asp 155. Glu 151 has an additional hydrogen bond with Gln 80. The calcium ion is surrounded by anomalous Fourier electron density contoured at 7.5 sigma colored in gold.
Figure 7. The second cytoplasmic domain of EpsF and a model of full-length EpsF dimer
(A) Alignment of the two cytoplasmic domains of T2SS homologs of EpsF. The two cytoplasmic domains from the same species as in Figure 1 were aligned. The upper block of sequences are sequences of the first cytoplasmic domain, the lower block of sequences represents the second cytoplasmic domain. Residue numbers above the sequences refer to the first cytoplasmic domain of EpsF from V. cholerae, while residue numbers below the sequences refer to the second cytoplasmic domain of EpsF from V. cholerae. Secondary structure assignment refers to that of cyto1-EpsF56-171. Identical residues are in red, similar residues according to the Risler matrix are in yellow. (B) Global model of a dimer formed by the cytoplasmic parts of two EpsF chains. The central part is identical to the cyto1-EpsF56-171 dimer (Figure 2). A second cytoplasmic domain is added with the same fold as the first domain on the basis of the high degree of sequence identity between the first and second EpsF domains (see text and Figure 7a). The orientation of the second domains with respect to the membrane is the same as of the first domains. The orientation of the second domains with respect to the first domains is arbitrary, but the two second domains are related by the same twofold axis relating the first cytoplasmic domains. Note that the N-terminal 70 residues, the two anti-parallel transmembrane helices TMH1 and TMH2, the 31-residue loop in the periplasm connecting these helices, and the C-terminal TMH3 plus the 17-residue tail in the periplasm are not included. The overall dimensions of this dimer are 81 by 55 Å parallel to the membrane, and 54 Å perpendicular to the inner membrane.
Figure 7. The second cytoplasmic domain of EpsF and a model of full-length EpsF dimer
(A) Alignment of the two cytoplasmic domains of T2SS homologs of EpsF. The two cytoplasmic domains from the same species as in Figure 1 were aligned. The upper block of sequences are sequences of the first cytoplasmic domain, the lower block of sequences represents the second cytoplasmic domain. Residue numbers above the sequences refer to the first cytoplasmic domain of EpsF from V. cholerae, while residue numbers below the sequences refer to the second cytoplasmic domain of EpsF from V. cholerae. Secondary structure assignment refers to that of cyto1-EpsF56-171. Identical residues are in red, similar residues according to the Risler matrix are in yellow. (B) Global model of a dimer formed by the cytoplasmic parts of two EpsF chains. The central part is identical to the cyto1-EpsF56-171 dimer (Figure 2). A second cytoplasmic domain is added with the same fold as the first domain on the basis of the high degree of sequence identity between the first and second EpsF domains (see text and Figure 7a). The orientation of the second domains with respect to the membrane is the same as of the first domains. The orientation of the second domains with respect to the first domains is arbitrary, but the two second domains are related by the same twofold axis relating the first cytoplasmic domains. Note that the N-terminal 70 residues, the two anti-parallel transmembrane helices TMH1 and TMH2, the 31-residue loop in the periplasm connecting these helices, and the C-terminal TMH3 plus the 17-residue tail in the periplasm are not included. The overall dimensions of this dimer are 81 by 55 Å parallel to the membrane, and 54 Å perpendicular to the inner membrane.
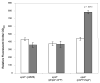
Figure 8. Fusion of GFP to the EpsF C-terminus results in a non-fluorescent protein
V. cholerae epsF mutant strains containing pMMB alone, or expressing either GFP-EpsF or EpsF-GFP were grown in M9 growth medium and carbenicillin without (white bars) and with (grey bars) IPTG at a final concentration of 20 μM to induce expression of the fusion proteins. Following growth, fluorescence was measured using the excitation and emisssion wavelengths 380 nm and 440 nm, respectively. Results from three independent experiments are presented as average relative fluorescence units ± standard error.
Similar articles
- Structural and functional studies of EpsC, a crucial component of the type 2 secretion system from Vibrio cholerae.
Korotkov KV, Krumm B, Bagdasarian M, Hol WG. Korotkov KV, et al. J Mol Biol. 2006 Oct 20;363(2):311-21. doi: 10.1016/j.jmb.2006.08.037. Epub 2006 Aug 18. J Mol Biol. 2006. PMID: 16978643 - The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae.
Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. Abendroth J, et al. J Mol Biol. 2005 May 13;348(4):845-55. doi: 10.1016/j.jmb.2005.02.061. J Mol Biol. 2005. PMID: 15843017 - Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL.
Lu C, Korotkov KV, Hol WGJ. Lu C, et al. J Struct Biol. 2014 Sep;187(3):223-235. doi: 10.1016/j.jsb.2014.07.006. Epub 2014 Aug 1. J Struct Biol. 2014. PMID: 25092625 Free PMC article. - Structure of the cytoplasmic domain of TcpE, the inner membrane core protein required for assembly of the Vibrio cholerae toxin-coregulated pilus.
Kolappan S, Craig L. Kolappan S, et al. Acta Crystallogr D Biol Crystallogr. 2013 Apr;69(Pt 4):513-9. doi: 10.1107/S0907444912050330. Epub 2013 Mar 9. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23519659
Cited by
- In through the Out Door: A Functional Virulence Factor Secretion System Is Necessary for Phage Infection in Ralstonia solanacearum.
Xavier ADS, de Melo AG, Hendrich CG, Tremblay DM, Rousseau GM, Plante PL, Forest KT, Alfenas-Zerbini P, Allen C, Moineau S. Xavier ADS, et al. mBio. 2022 Dec 20;13(6):e0147522. doi: 10.1128/mbio.01475-22. Epub 2022 Oct 31. mBio. 2022. PMID: 36314808 Free PMC article. - Structural insights into the Type II secretion nanomachine.
McLaughlin LS, Haft RJ, Forest KT. McLaughlin LS, et al. Curr Opin Struct Biol. 2012 Apr;22(2):208-16. doi: 10.1016/j.sbi.2012.02.005. Epub 2012 Mar 16. Curr Opin Struct Biol. 2012. PMID: 22425326 Free PMC article. Review. - Electron Cryotomography of Bacterial Secretion Systems.
Oikonomou CM, Jensen GJ. Oikonomou CM, et al. Microbiol Spectr. 2019 Mar;7(2):10.1128/microbiolspec.psib-0019-2018. doi: 10.1128/microbiolspec.PSIB-0019-2018. Microbiol Spectr. 2019. PMID: 30953431 Free PMC article. - Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
Douzi B, Trinh NTT, Michel-Souzy S, Desmyter A, Ball G, Barbier P, Kosta A, Durand E, Forest KT, Cambillau C, Roussel A, Voulhoux R. Douzi B, et al. mBio. 2017 Oct 17;8(5):e01185-17. doi: 10.1128/mBio.01185-17. mBio. 2017. PMID: 29042493 Free PMC article. - Secretion systems in Gram-negative bacteria: structural and mechanistic insights.
Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. Costa TR, et al. Nat Rev Microbiol. 2015 Jun;13(6):343-59. doi: 10.1038/nrmicro3456. Nat Rev Microbiol. 2015. PMID: 25978706 Review.
References
- Abendroth J, Bagdasarian M, Sandkvist M, Hol WGJ. The structure of the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae: An unusual member of the actin-like ATPase superfamily. Journal of Molecular Biology. 2004a;344:619–633. - PubMed
- Abendroth J, Rice AE, McLuskey K, Bagdasarian M, Hol WGJ. The crystal structure of the periplasmic domain of the type II secretion system protein EpsM from Vibrio cholerae: The simplest version of the ferredoxin fold. Journal of Molecular Biology. 2004b;338:585–596. - PubMed
- Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WGJ. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. Journal of Molecular Biology. 2005;348:845–855. - PubMed
- Arts J, de Groot A, Ball G, Durand E, El Khattabi M, Filloux A, Tommassen J, Koster M. Interaction domains in the Pseudomonas aeruginosa type II secretory apparatus component XcpS (GspF) Microbiology-Uk. 2007;153:1582–1592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI34501/AI/NIAID NIH HHS/United States
- AI49294/AI/NIAID NIH HHS/United States
- R01 AI049294/AI/NIAID NIH HHS/United States
- R01 AI034501/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R56 AI034501/AI/NIAID NIH HHS/United States
- R56 AI049294/AI/NIAID NIH HHS/United States
- R01 AI034501-15/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases