Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens - PubMed (original) (raw)
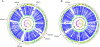
Figure 1. Schematic circular diagrams of the _Se_4047 (A) and _Sz_H70 genomes (B).
Key for the circular diagrams (outside to inside): scale (in Mb); annotated CDSs colored according to predicted function represented on a pair of concentric circles, representing both coding strands; orthologue matches shared with the Streptococcal species, _Se_4047 or _Sz_H70, Sz_MGCS10565, S. uberis 0140J, S. pyogenes Manfredo, S. mutans UA159, S. gordonii Challis CH1, S. sanguinis SK36, S. pneumoniae TIGR4, S. agalactiae NEM316, S. suis P1/7, S. thermophilus CNRZ1066, blue; orthologue matches shared with Lactococcus lactis subspecies lactis, green; G+C% content plot; G+C deviation plot (>0%, olive, <0%, purple). Color coding for CDS functions: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface-associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage and IS elements; grey, miscellaneous. The positions of the four prophage and two ICE_Se present in the Se_4047 genome, and two ICE_Sz in the _Sz_H70 genome, are indicated.
Figure 2. Pairwise comparison of the chromosomes of _Se_4047 and _Sz_H70 using ACT.
The sequences have been aligned from the predicted replication origins (oriC; right). The colored bars separating each genome (red and blue) represent similarity matches identified by reciprocal TBLASTX analysis , with a score cutoff of 100. Red lines link matches in the same orientation; blue lines link matches in the reverse orientation. The prophage (pink) and ICE (purple) are highlighted as colored boxes.
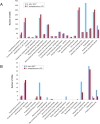
Figure 3. Distribution of CDSs belonging to different functional classes in the _Se_4047 and _Sz_H70 genomes.
(A) Functional CDSs and pseudogenes of _Se_4047 and _Sz_H70. (B) Partially deleted or pseudogenes in the _Se_4047 and _Sz_H70 genomes.
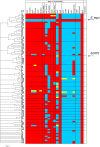
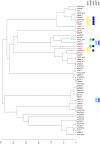
Figure 4. ClonalFrame analysis of MLST alleles of 26 S. equi and 140 S. zooepidemicus isolates and its relationship with the prevalence of selected differences between the _Se_4047 and _Sz_H70 genomes.
Genes examined were lacE, rbsD, sorD, SZO06680 (encoding a putative hyaluronate lyase and specific to the 4 bp missing from SEQ1479), srtC, srtD, SZO08560 (encoding a Listeria-Bacteroides repeat domain containing surface-anchored protein), esaA, SZO14370 (within the CRISPR locus), slaA, slaB, seeL, seeM, seeH, seeI, eqbE (within the equibactin locus), SEQ0235 (encoding Se18.9), and gyrA. Functional assays determined the ability of different isolates to ferment lactose, ribose, and sorbitol and to induce mitogenic responses in equine peripheral blood mononuclear cells. The number of isolates representing each ST is indicated. STs where all isolates contained the gene or possessed functional activity are shown in red, STs where all isolates lacked the gene or functionality are shown in blue, and STs containing some isolates containing the gene or functionality and some that did not are colored in yellow. The position of S. equi isolates and _Sz_H70 are indicated. _Sz_MGCS10565 is a single locus variant of ST-10 (ST-72; not shown), and had an identical gene prevalence profile to the ST-10 isolates based on in silico analysis of its genome sequence .
Figure 5. Diagram of the SZO08560 invertible promoter in _Sz_H70.
The promoter region of SZO08560 (−170 bp to −55 bp) is bordered by GTAGACTTTA and TAAAGTCTAC inverted repeats that invert to switch transcription from forward to reverse orientation.
Figure 6. Clustering of _Se_4047 prophage with S. pyogenes prophage.
UPGMA tree generated from tribeMCL clustering of CDSs from _Se_4047 prophage (highlighted in red) and S. pyogenes prophage. S. pyogenes prophage used in the clustering were: Manfredo (ϕMan.1, ϕMan.2, ϕMan.3, ϕMan.4, and ϕMan.5), SSI-1 (SPsP1, SPsP2, SPsP3, SPsP4, and SPsP5), SF370 (370.1, 370.2, 370.3, and 370.4), MGAS315 (ϕ315.1, ϕ315.2, ϕ315.3, ϕ315.4, ϕ315.5, and ϕ315.6), MGAS8232 (ϕspeA, ϕspeC, ϕspeL/M, ϕ370.3-like, and ϕsda), MGAS10394 (ϕ10394.1, ϕ10394.2, ϕ10394.3, ϕ10394.4, ϕ10394.5, ϕ10394.6, ϕ10394.7, and ϕ10394.8), MGAS6180 (ϕ6180.1, ϕ6180.2, ϕ6180.3, and ϕ6180.4), MGAS5005 (ϕ5005.1, ϕ5005.2, and ϕ5005.3), MGAS2096 (ϕ2096.1 and ϕ2096.2), MGAS9429 (ϕ9429.1, ϕ9429.2, and ϕ9429.3), MGAS10270 (ϕ10270.1, ϕ10270.2, ϕ10270.3, 10270.4, and 10270.5), and MGAS10750 (ϕ10270.1, ϕ10270.2, ϕ10270.3, and ϕ10270.4). The distribution of homologues to virulence cargo of _Se_4047 prophage are indicated on the right hand side. CDSs belonging to the same homology groups defined using TribeMCL with a cutoff of 1e−5 are indicated by colored blocks: slaA (yellow), seeM (green), seeL (dark blue), seeH (light blue), and seeI (pink).
Figure 7. Pairwise comparison of _Se_4047 ϕSeq.4 and ϕMan.3 from S. pyogenes Manfredo displayed using ACT.
The red bars separating each sequence represent similarity matches identified by TBLASTX analysis. The locations of seeI, seeH, speI, and speH are indicated.
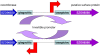
Figure 8. Summary of functional loss and gene gain by S. equi.
Gene loss (blue): (1) _Se_4047 has lost the ability to ferment lactose, sorbitol, and ribose, which may reduce its ability to colonize the mucosal surface. (2) Hyaluronate lyase activity is predicted to be reduced in _Se_4047, which could decrease its ability to invade tissue and provide an explanation for increased levels of hyaluronate capsule. Increased levels of capsule may enhance resistance to phagocytosis, but could also reduce adhesion to the mucosal surface. (3) Truncation of fne and Shr in _Se_4047 and subsequent synthesis of secreted fibronectin products may decrease the adhesive properties of _Se_4047 and interfere with fibronectin-dependent attachment mechanisms of competing pathogens. (4) Loss of function of the tetR regulator may lead to constitutive production of longer collagen-binding pili by S. equi. (5) The putative SZO18310 pilus locus of _Sz_H70 has been deleted from the _Se_4047 genome. (6) _Se_4047 has lost a Listeria-Bacteroides repeat domain containing surface-anchored protein. Gene gain (red): (7) The acquisition of prophage plays an important evolutionary role through integration of cargo genes. (8) Recirculation and secretion of the integrated ϕSeq1 may kill susceptible competing bacteria such as S. zooepidemicus. (9) ϕSeq2 contains a gene encoding a phospholipase A2 (SlaA) that may enhance virulence. (10) ϕSeq3 and ϕSeq4 encode superantigens SeeH, SeeI, SeeL, and SeeM that target the equine immune system (11). (12) The absence of prophage in S. zooepidemicus may be explained by the presence of CRISPR arrays and competence proteins that confer resistance to circulating phage and maintain genome integrity. (13) The ICE_Se_2 locus may enhance iron acquisition in _Se_4047 through the production of a potential siderophore, equibactin. (14) Se18.9 binds Factor H and interferes with complement activation.