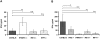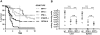Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes - PubMed (original) (raw)
. 2009 Mar;5(3):e1000355.
doi: 10.1371/journal.ppat.1000355. Epub 2009 Mar 27.
Renate Kastner, Elisabeth Kernbauer, Andreas Pilz, Sandra Westermayer, Benjamin Reutterer, Didier Soulat, Gabriele Stengl, Claus Vogl, Theresa Frenz, Zoe Waibler, Tadatsugu Taniguchi, Thomas Rülicke, Ulrich Kalinke, Mathias Müller, Thomas Decker
Affiliations
- PMID: 19325882
- PMCID: PMC2654726
- DOI: 10.1371/journal.ppat.1000355
Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes
Silvia Stockinger et al. PLoS Pathog. 2009 Mar.
Abstract
Production of type I interferons (IFN-I, mainly IFNalpha and IFNbeta) is a hallmark of innate immune responses to all classes of pathogens. When viral infection spreads to lymphoid organs, the majority of systemic IFN-I is produced by a specialized "interferon-producing cell" (IPC) that has been shown to belong to the lineage of plasmacytoid dendritic cells (pDC). It is unclear whether production of systemic IFN-I is generally attributable to pDC irrespective of the nature of the infecting pathogen. We have addressed this question by studying infections of mice with the intracellular bacterium Listeria monocytogenes. Protective innate immunity against this pathogen is weakened by IFN-I activity. In mice infected with L. monocytogenes, systemic IFN-I was amplified via IFN-beta, the IFN-I receptor (IFNAR), and transcription factor interferon regulatory factor 7 (IRF7), a molecular circuitry usually characteristic of non-pDC producers. Synthesis of serum IFN-I did not require TLR9. In contrast, in vitro-differentiated pDC infected with L. monocytogenes needed TLR9 to transcribe IFN-I mRNA. Consistent with the assumption that pDC are not the producers of systemic IFN-I, conditional ablation of the IFN-I receptor in mice showed that most systemic IFN-I is produced by myeloid cells. Furthermore, results obtained with FACS-purified splenic cell populations from infected mice confirmed the assumption that a cell type with surface antigens characteristic of macrophages and not of pDC is responsible for bulk IFN-I synthesis. The amount of IFN-I produced in the investigated mouse lines was inversely correlated to the resistance to lethal infection. Based on these data, we propose that the engagement of pDC, the mode of IFN-I mobilization, as well as the shaping of the antimicrobial innate immune response by IFN-I differ between intracellular pathogens.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Figure 1. Serum levels of IFN-I in mice deficient in components of the IFN-I signaling pathway.
Mice of the indicated genotypes were injected intraperitoneally (i.p.) with 5×106 L. monocytogenes (C57BL/6 n = 25, IFNβ−/− n = 25, IFNAR1−/− n = 24, IRF3−/− n = 25, IRF7−/− n = 20) or PBS as a control (data not shown). After 24 h, serum was collected and ELISAs for IFNβ (A) and IFNα (B) were performed. The data presented here are a summary of several individual experiments with groups of four to five infected mice per genotype. Data representing IFNβ (A) and IFNα (B) concentrations were log-transformed (after adding one) to achieve approximate normality. Linear models with genotype and experiment as fixed effects were fitted using SPSS. Means plus/minus standard errors of wt and mutant genotypes are plotted (after back-transformation). Significant values are indicated by: n.s. not significant p>0.05, * p≤0.05, ** p≤0.01, *** p≤0.001.
Figure 2. Prerequisites for IFN-I production in infected spleens and _in vitro_–generated BMM and mDCs.
Mice of the indicated genotypes were injected i.p. with 5×106 L. monocytogenes or PBS as a control. After 24 h of infection, mice were killed and spleens and livers isolated. (A) RNA was extracted from spleens of four infected mice per genotype (1–4) or from mice injected with PBS (−), and RT-PCR for the indicated genes was performed. As a normalization control, the housekeeping gene L32 was measured. (B) Summary of experiments performed with mice of the indicated genotypes. In total, 20 C57BL/6, 20 IFNβ−/−, 19 IFNAR1−/−, 20 IRF3−/−, and 15 IRF7−/− mice were analysed for IFN-I expression and expression of Gbp2. The graph shows the percentage of mice of the respective genotypes showing detectable IFNα (black) or IFNβ (grey) expression after infection with 5×106 L. monocytogenes for 24 h. (C–F) BMM (C,D) or mDC (E,F) of the indicated genotypes were infected with L. monocytogenes at a MOI of 10. At the indicated time points, total RNA was prepared. The isolated RNA was reverse-transcribed and induction of the IFNβ (C,E) or IFNα genes (D,F) was measured by real-time PCR. For normalization to a housekeeping gene, GAPDH was measured.
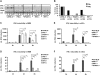
Figure 3. Myeloid cells strongly contribute to IFN-I production.
(A) C57BL/6 wt (n = 13), IFNAR1−/− (n = 4), and mice with LysM-Cre-mediated deletion of IFNAR1 (LysMCre-IFNAR1fl/fl, n = 9) were infected with 5×106 L. monocytogenes, and survival was monitored for ten days. (B) Mice of the indicated genotypes were infected with 5×106 L. monocytogenes (seven mice per group) or injected with PBS (two mice per group). After 24 h, serum was collected and ELISA was performed for IFNβ (data not shown) and IFNα levels. Data representing IFNα concentrations were log-transformed to achieve approximate normality. Linear models with genotype as fixed effect were fitted using SPSS. Means plus/minus standard errors of wt and mutant genotypes are plotted (after back-transformation). Significant values are indicated by ** p≤0.01, *** p≤0.001. (C) Mice of the indicated genotypes were infected with 5×106 L. monocytogenes (1–3) or injected with PBS (−). After 24 h and 48 h, spleens were isolated and RNA was extracted. The RNA was reverse-transcribed and subjected to PCR for the indicated genes. As a normalization control, the housekeeping gene L32 was measured.
Figure 4. Prerequisites for the increase in resistance to lethal infection caused by IFN-I.
(A) Mice of the indicated genotypes were infected with 5×106 L. monocytogenes, and survival was monitored for ten days. The data presented here are a summary of several individual experiments with groups of 4–6 mice per genotype and treatment. Total number of mice analysed are: C57BL/6 wt n = 34, IFNAR1−/− n = 23, IFNβ−/− n = 23, IRF3−/− n = 24, IRF7−/− n = 29. (B) Groups of ten (C57BL/6 wt and IFNβ−/−) or eight (IRF7−/−) mice were infected with 1×106 L. monocytogenes. After three days, mice were killed and the L. monocytogenes titre was determined in the liver and spleen and presented as CFU. Data were log-transformed to achieve approximate normality. Linear models with genotype as fixed effect were fitted using SPSS. Significant values are indicated by: n.s. not significant p>0.05, *** p≤0.001.
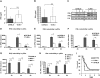
Figure 5. The role of TLR9 in mice and cells infected with L. monocytogenes.
(A–C) C57BL/6 wt or TLR9−/− mice were injected i.p. with 5×106 L. monocytogenes (four mice per genotype) or PBS as a control (data not shown). After 24 h of infection, serum was collected and ELISAs for IFNβ (A) and IFNα (B) were performed. Data representing IFNβ (A) and IFNα (B) concentrations were log-transformed (after adding one) to achieve approximate normality. Linear models with genotype as fixed effect were fitted using SPSS. Means plus/minus standard errors of wt and mutant genotypes are plotted (after back-transformation), n.s.: not significant (p>0.05). (C) After 24 h of infection, mice were killed and spleens isolated. RNA was extracted from spleens of three infected mice per genotype (1–3) or from mice injected with PBS (−), and RT-PCR for the indicated genes was performed. As a normalization control, the housekeeping gene L32 was measured. (D–H) pDCs of the indicated genotypes were infected with L. monocytogenes at a MOI of 10. At the indicated time points, total RNA was prepared. The isolated RNA was reverse-transcribed, and induction of the IFNβ (D,E,G) or IFNα (F,H) genes was measured by real-time PCR. For normalization to a housekeeping gene, GAPDH was measured. (I) C57BL/6 wt or TLR9−/− mice (13 mice per genotype, data represent a summary of two experiments) were injected i.p. with 5×106 L. monocytogenes. Survival was monitored for ten days.
Figure 6. Macrophages and not pDC are the IFN-I–producing cells after L. monocytogenes infection.
C57Bl/6 wt mice (n = 15) were infected with 5×106 L. monocytogenes. After 24 h, mice were killed, splenic cells were isolated, labelled with antibodies against CD11b (FITC-conjugated), CD11c (APC), PDCA1 (PE-Cy5), and B220 (PE-Cy7), and subjected to FACS sorting. RNA of the isolated cell populations was extracted, reverse-transcribed, and subjected to PCR for the indicated genes. Unsorted cells and whole spleen extracts from infected spleens (crude extract) as well as uninfected spleens were used as controls. As a positive control for the PCR reactions, Raw264.7 macrophages were infected (+) or not (−) with L. monocytogenes for 8 h at a MOI of 10. As a negative control, H2O was used instead of template cDNA. As a normalization control, the housekeeping gene L32 was measured.
Similar articles
- Type I IFN Does Not Promote Susceptibility to Foodborne Listeria monocytogenes.
Pitts MG, Myers-Morales T, D'Orazio SE. Pitts MG, et al. J Immunol. 2016 Apr 1;196(7):3109-16. doi: 10.4049/jimmunol.1502192. Epub 2016 Feb 19. J Immunol. 2016. PMID: 26895837 Free PMC article. - Type I IFN induction in response to Listeria monocytogenes in human macrophages: evidence for a differential activation of IFN regulatory factor 3 (IRF3).
Reimer T, Schweizer M, Jungi TW. Reimer T, et al. J Immunol. 2007 Jul 15;179(2):1166-77. doi: 10.4049/jimmunol.179.2.1166. J Immunol. 2007. PMID: 17617610 - Conditional Stat1 ablation reveals the importance of interferon signaling for immunity to Listeria monocytogenes infection.
Kernbauer E, Maier V, Stoiber D, Strobl B, Schneckenleithner C, Sexl V, Reichart U, Reizis B, Kalinke U, Jamieson A, Müller M, Decker T. Kernbauer E, et al. PLoS Pathog. 2012;8(6):e1002763. doi: 10.1371/journal.ppat.1002763. Epub 2012 Jun 14. PLoS Pathog. 2012. PMID: 22719255 Free PMC article. - Novel functions of type I interferons revealed by infection studies with Listeria monocytogenes.
Stockinger S, Decker T. Stockinger S, et al. Immunobiology. 2008;213(9-10):889-97. doi: 10.1016/j.imbio.2008.07.020. Epub 2008 Sep 2. Immunobiology. 2008. PMID: 18926303 Review. - Antagonistic crosstalk between type I and II interferons and increased host susceptibility to bacterial infections.
Rayamajhi M, Humann J, Kearney S, Hill KK, Lenz LL. Rayamajhi M, et al. Virulence. 2010 Sep-Oct;1(5):418-22. doi: 10.4161/viru.1.5.12787. Virulence. 2010. PMID: 21178482 Free PMC article. Review.
Cited by
- Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse.
Carrero JA, Calderon B, Towfic F, Artyomov MN, Unanue ER. Carrero JA, et al. PLoS One. 2013;8(3):e59701. doi: 10.1371/journal.pone.0059701. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555752 Free PMC article. - Plasmacytoid dendritic cells in the eye.
Jamali A, Kenyon B, Ortiz G, Abou-Slaybi A, Sendra VG, Harris DL, Hamrah P. Jamali A, et al. Prog Retin Eye Res. 2021 Jan;80:100877. doi: 10.1016/j.preteyeres.2020.100877. Epub 2020 Jul 24. Prog Retin Eye Res. 2021. PMID: 32717378 Free PMC article. Review. - STING/MPYS mediates host defense against Listeria monocytogenes infection by regulating Ly6C(hi) monocyte migration.
Jin L, Getahun A, Knowles HM, Mogan J, Akerlund LJ, Packard TA, Perraud AL, Cambier JC. Jin L, et al. J Immunol. 2013 Mar 15;190(6):2835-43. doi: 10.4049/jimmunol.1201788. Epub 2013 Feb 1. J Immunol. 2013. PMID: 23378430 Free PMC article. - The bacterial pathogen Listeria monocytogenes and the interferon family: type I, type II and type III interferons.
Dussurget O, Bierne H, Cossart P. Dussurget O, et al. Front Cell Infect Microbiol. 2014 Apr 28;4:50. doi: 10.3389/fcimb.2014.00050. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24809023 Free PMC article. Review. - Listeria monocytogenes induces an interferon-enhanced activation of the integrated stress response that is detrimental for resolution of infection in mice.
Valderrama C, Clark A, Urano F, Unanue ER, Carrero JA. Valderrama C, et al. Eur J Immunol. 2017 May;47(5):830-840. doi: 10.1002/eji.201646856. Epub 2017 Apr 13. Eur J Immunol. 2017. PMID: 28267207 Free PMC article.
References
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. - PubMed
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. - PubMed
- Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. - PubMed
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. - PubMed
- Jefferies CA, Fitzgerald KA. Interferon gene regulation: not all roads lead to Tolls. Trends Mol Med. 2005;11:403–411. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical