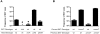A toxin-antitoxin system promotes the maintenance of an integrative conjugative element - PubMed (original) (raw)
A toxin-antitoxin system promotes the maintenance of an integrative conjugative element
Rachel A F Wozniak et al. PLoS Genet. 2009 Mar.
Abstract
SXT is an integrative and conjugative element (ICE) that confers resistance to multiple antibiotics upon many clinical isolates of Vibrio cholerae. In most cells, this approximately 100 Kb element is integrated into the host genome in a site-specific fashion; however, SXT can excise to form an extrachromosomal circle that is thought to be the substrate for conjugative transfer. Daughter cells lacking SXT can theoretically arise if cell division occurs prior to the element's reintegration. Even though approximately 2% of SXT-bearing cells contain the excised form of the ICE, cells that have lost the element have not been detected. Here, using a positive selection-based system, SXT loss was detected rarely at a frequency of approximately 1 x 10(-7). As expected, excision appears necessary for loss, and factors influencing the frequency of excision altered the frequency of SXT loss. We screened the entire 100 kb SXT genome and identified two genes within SXT, now designated mosA and mosT (for maintenance of SXT Antitoxin and Toxin), that promote SXT stability. These two genes, which lack similarity to any previously characterized genes, encode a novel toxin-antitoxin pair; expression of mosT greatly impaired cell growth and mosA expression ameliorated MosT toxicity. Factors that promote SXT excision upregulate mosAT expression. Thus, when the element is extrachromosomal and vulnerable to loss, SXT activates a TA module to minimize the formation of SXT-free cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
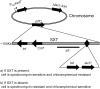
Figure 1. Schematic of a positively selectable reporter of SXT loss.
SXT containing lacIQ was introduced into a lacI E. coli host containing Plac_-aad7_ (a spectinomycin-resistance gene). If SXT (and lacIQ) is lost, the cells become SpecR. The black diamond represents the site of insertion for a fragment containing lacIQ (pFRTIq), thick black arrows represent ORFs, and the thin black line indicates the approximate location of the Δ9 deletion. Abbreviations: tet = tetracycline resistance gene, kan = kanamycin resistance gene, int = integrase, prfC = site of SXT insertion. Figure not to scale.
Figure 2. Influence of genetic factors on the frequency of SXT loss.
The frequency of SXT loss was calculated as the ratio of SpecRCmS cfu / total cfu after 15 hour of growth. A) Factors that influence the frequency of SXT excision alter the frequency of SXT loss. The black diamonds indicate that the result is statistically different (p<0.05) than the result shown in the first column for the wild type reporter. The * signifies that the result was below the limit of detection which was ∼1×10−8. pXis and pXis-R harbor arabinose-inducible xis or its reverse complement respectively. B) SXT loss is not accompanied by heritable changes that influence maintenance of SXT. The ‘current SXT genotype’ refers to the ICE introduced into the host that lost the ICE designated as ‘previous SXT genotype’.
Figure 3. Genetic analysis of loss of mosT SXT.
A) Schematic of the region deleted from Δ9 SXT. Black arrows represent ORFs, thin black arrows represent the deletions studied below. B) Frequency of loss of the indicated mutant SXT. C) Influence of mosT expression in trans on the stability of mosT SXT. All cultures were grown in the presence of 0.02% arabinose for 15 hr. Black diamond represents a statistically significant (p<.05) result compared to loss in the presence of pBAD33. D) Loss of mosT SXT is influenced by factors affecting SXT excision. All cultures were grown for 15 hr except as noted. Black diamonds represent a statistically significant (p<0.05) result compared to the mosT SXT grown for 15 hr. The * signifies that the result was below the limit of detection which was ∼1×10−8. aLoss was calculated following 3 hours of growth in an early log phase culture.
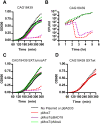
Figure 4. MosT inhibits E. coli growth and its toxicity can be neutralized by MosA.
Growth kinetics of E. coli strains CAG18439 (A and B), CAG18439 containing mosAT SXT (C) and CAG18439 containing wild type SXT (D). These strains, which harbored arabinose-inducible mosT and mosA, (pMosT or pMosA respectively) or control vectors, (pBAD33 and pBAD18), were grown in either 0.2% glucose (solid lines) or 0.02% arabinose (dashed lines).
Figure 5. 5′RACE analysis of mosA transcription start site.
A) Schematic of mosA transcription start site based on the 5′ RACE results shown in B). The +1 refers to the start of transcription as defined by the DNA sequence of the 5′RACE product obtained using primer A (small black arrow). The length of the PCR product is indicated. Black arrows represent ORFs as predicted using bioinformatics; the previously annotated s052 start codon is indicated. B) Shows a 1% agarose gel of the product of 5′RACE reaction using primer A.
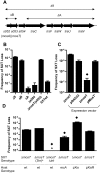
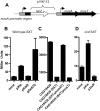
Figure 6. Influence of Xis and SetCD on mosA expression.
A) Schematic of chromosomal mosA:: lacZ transcription reporter within the mosAT locus. Thick black arrows represent ORFs as predicted by bioinformatics and the thin arrow represents the predicted location of the mosA promoter. B, D) β-galactosidase activities of the chromosomal mosA::lacZ fusion in CAG18439 containing wt SXT (B) or in CAG18439 containing Δint SXT (D) along with the indicating expression vectors. C) β-galactasidase activities from a plasmid-borne mosA-lacZ fusion (in pPmosA) in the indicated strains. All β-galactasidase measurements were conducted on 15 hr cultures and the results shown are the means and standard deviation from at least 9 independent cultures.
Similar articles
- The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2.
Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D. Dziewit L, et al. J Bacteriol. 2007 Mar;189(5):1983-97. doi: 10.1128/JB.01610-06. Epub 2006 Dec 8. J Bacteriol. 2007. PMID: 17158670 Free PMC article. - Control of SXT integration and excision.
Burrus V, Waldor MK. Burrus V, et al. J Bacteriol. 2003 Sep;185(17):5045-54. doi: 10.1128/JB.185.17.5045-5054.2003. J Bacteriol. 2003. PMID: 12923077 Free PMC article. - Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae.
Beaber JW, Hochhut B, Waldor MK. Beaber JW, et al. J Bacteriol. 2002 Aug;184(15):4259-69. doi: 10.1128/JB.184.15.4259-4269.2002. J Bacteriol. 2002. PMID: 12107144 Free PMC article. - The current ICE age: biology and evolution of SXT-related integrating conjugative elements.
Burrus V, Marrero J, Waldor MK. Burrus V, et al. Plasmid. 2006 May;55(3):173-83. doi: 10.1016/j.plasmid.2006.01.001. Epub 2006 Mar 13. Plasmid. 2006. PMID: 16530834 Review. - [Bacterial SXT/R391 family from integrating conjugative elements--a review].
Luo P, Hu C. Luo P, et al. Wei Sheng Wu Xue Bao. 2014 May 4;54(5):471-9. Wei Sheng Wu Xue Bao. 2014. PMID: 25199245 Review. Chinese.
Cited by
- Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs.
Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. Wozniak RA, et al. PLoS Genet. 2009 Dec;5(12):e1000786. doi: 10.1371/journal.pgen.1000786. Epub 2009 Dec 24. PLoS Genet. 2009. PMID: 20041216 Free PMC article. - A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism.
Dy RL, Przybilski R, Semeijn K, Salmond GP, Fineran PC. Dy RL, et al. Nucleic Acids Res. 2014 Apr;42(7):4590-605. doi: 10.1093/nar/gkt1419. Epub 2014 Jan 24. Nucleic Acids Res. 2014. PMID: 24465005 Free PMC article. - Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans.
Hu Y, Zhu Y, Ma Y, Liu F, Lu N, Yang X, Luan C, Yi Y, Zhu B. Hu Y, et al. Antimicrob Agents Chemother. 2015 Feb;59(2):1152-61. doi: 10.1128/AAC.04260-14. Epub 2014 Dec 8. Antimicrob Agents Chemother. 2015. PMID: 25487802 Free PMC article. - Integrative and Conjugative Elements (ICEs): What They Do and How They Work.
Johnson CM, Grossman AD. Johnson CM, et al. Annu Rev Genet. 2015;49:577-601. doi: 10.1146/annurev-genet-112414-055018. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473380 Free PMC article. Review. - Interaction of Type IV Toxin/Antitoxin Systems in Cryptic Prophages of Escherichia coli K-12.
Wen Z, Wang P, Sun C, Guo Y, Wang X. Wen Z, et al. Toxins (Basel). 2017 Mar 1;9(3):77. doi: 10.3390/toxins9030077. Toxins (Basel). 2017. PMID: 28257056 Free PMC article.
References
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. - PubMed
- Churchward G. Back to the future: the new ICE age. Mol Microbiol. 2008;70:554–556. - PubMed
- Pavlovic G, Burrus V, Gintz B, Decaris B, Guedon G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology. 2004;150:759–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI042347/AI/NIAID NIH HHS/United States
- R37 AI042347/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- AI-042347/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials