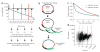Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming - PubMed (original) (raw)
doi: 10.1038/nbt.1530. Epub 2009 Mar 29.
Robert Shoemaker, Bin Xie, Athurva Gore, Emily M LeProust, Jessica Antosiewicz-Bourget, Dieter Egli, Nimet Maherali, In-Hyun Park, Junying Yu, George Q Daley, Kevin Eggan, Konrad Hochedlinger, James Thomson, Wei Wang, Yuan Gao, Kun Zhang
Affiliations
- PMID: 19330000
- PMCID: PMC2715272
- DOI: 10.1038/nbt.1530
Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming
Jie Deng et al. Nat Biotechnol. 2009 Apr.
Abstract
Current DNA methylation assays are limited in the flexibility and efficiency of characterizing a large number of genomic targets. We report a method to specifically capture an arbitrary subset of genomic targets for single-molecule bisulfite sequencing for digital quantification of DNA methylation at single-nucleotide resolution. A set of ~30,000 padlock probes was designed to assess methylation of ~66,000 CpG sites within 2,020 CpG islands on human chromosome 12, chromosome 20, and 34 selected regions. To investigate epigenetic differences associated with dedifferentiation, we compared methylation in three human fibroblast lines and eight human pluripotent stem cell lines. Chromosome-wide methylation patterns were similar among all lines studied, but cytosine methylation was slightly more prevalent in the pluripotent cells than in the fibroblasts. Induced pluripotent stem (iPS) cells appeared to display more methylation than embryonic stem cells. We found 288 regions methylated differently in fibroblasts and pluripotent cells. This targeted approach should be particularly useful for analyzing DNA methylation in large genomes.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/
Figures
Figure 1
Targeted bisulfite sequencing with padlock probes. (a) Each padlock probe has a common linker sequence flanked by two target-specific capturing arms (H1 and H2). H1 and H2 are melting temperature–normalized, and a spacer sequence is included to normalize probe lengths. The linker sequence contains priming sites (AP1 and AP2) for universal primers, two _Mme_I sites and a central _Alu_I recognition site. (b) A CpG island (or other target regions) is covered by multiple padlock probes targeting partially overlapped regions on alternating strands. (c) A library of padlock probes is annealed to bisulfite-converted genomic DNA (black) and the 3′ ends are extended and ligated with the 5′ end. After removal of linear DNAs with exonucleases, all circularized padlock probes are PCR-amplified using a pair of common primers. (d) To generate a shotgun sequencing library, amplicons were reamplified in the presence of dUTP, digested with _Mme_I, and then with USER and S1 nuclease. Digested amplicons are end repaired and ligated with Solexa sequencing adaptors (green). Ligated products are then selected by size and amplified by PCR to generate the shotgun sequencing library. (e) Gel electrophoresis analysis of the padlock-captured products from two independent capturing reactions (1 and 2) and a no-template control. Expected amplicon size is in the range of 344–394 bp, which includes capturing targets (175–225 bp), capturing arms (58 bp) and amplification primers (111 bp). NTC, no-template control.
Figure 2
Normalization of padlock-capturing efficiency. (a) The ‘subsetting’ strategy. The 30,000 probes were divided into four sets (5 k, 10 k, 10 k, 5 k). The three less efficient sets were resynthesized. We reused the original 30,000-probe set because it was dominated by the most efficient 5,000 probes. (b) The ‘suppressor oligo’ strategy. (c) Distribution of normalized abundance for all captured targets with one 30,000-probe set and with four probe sets. The _x_-axis is the normalized abundance of each captured target, which is calculated by dividing the counts of the target by the average counts of all targets. The _y_-axis is the fraction of probes with the coverage equal to or greater than the normalized coverage. (d) Comparison of relative abundance for each target before and after normalization. The green vertical dash lines indicate the clear separation of four subsets of targets, as well as the fifth set normalized with the suppressor oligos.
Figure 3
Patterns of CpG methylation in fibroblasts and pluripotent cells. (a,b) Patterns of CpG island methylation on chromosomes 12 and 20 in twelve samples. Red indicates highly methylated CpG, and green indicates weakly methylated CpG. (c) Correlation between DNA methylation and gene expression. Each circle or square represents a 500-bp window. The windows in which average methylation levels are significantly correlated (P < 0.01) with gene expression are highlighted as orange circles. Gray squares indicate that the correlation is not significant. (d) Clustering of 26 selected genes based on the distribution of CpG methylation in the 4-kb TSS flanking regions. The 20 columns per cell line represent the fraction of probes within the TSS region where each probe exhibits a methylation frequency such that column header value ≤ methylation frequency < column header value + 0.05. For example, unmethylated TSS regions will have the highest probe fraction in the lowest value-labeled (left) column. Roughly half of the genes (14) were close to completely unmethylated in all cell types. Five genes (OCT4, ZIC3, NANOG, UTF1, DNMT3B) that show cell-type specific changes in methylation were grouped as a cluster. (e) Methylation pattern in the TSS flanking regions of OCT4. Two differentially methylated regions were marked by purple rectangles. (f) Hierarchical clustering of pluripotent cells and fibroblasts. The dissimilarity matrix was calculated based on Pearson correlation coefficients.
Comment in
- Locking in on the human methylome.
Berman BP, Weisenberger DJ, Laird PW. Berman BP, et al. Nat Biotechnol. 2009 Apr;27(4):341-2. doi: 10.1038/nbt0409-341. Nat Biotechnol. 2009. PMID: 19352369 No abstract available.
Similar articles
- Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells.
Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Ball MP, et al. Nat Biotechnol. 2009 Apr;27(4):361-8. doi: 10.1038/nbt.1533. Epub 2009 Mar 29. Nat Biotechnol. 2009. PMID: 19329998 Free PMC article. - Large-Scale Targeted DNA Methylation Analysis Using Bisulfite Padlock Probes.
Diep D, Plongthongkum N, Zhang K. Diep D, et al. Methods Mol Biol. 2018;1708:365-382. doi: 10.1007/978-1-4939-7481-8_19. Methods Mol Biol. 2018. PMID: 29224154 - Methodological aspects of whole-genome bisulfite sequencing analysis.
Adusumalli S, Mohd Omar MF, Soong R, Benoukraf T. Adusumalli S, et al. Brief Bioinform. 2015 May;16(3):369-79. doi: 10.1093/bib/bbu016. Epub 2014 May 27. Brief Bioinform. 2015. PMID: 24867940 Review. - Bisulfite Sequencing for DNA Methylation Analysis of Primary Muscle Stem Cells.
Miyata K, Naito M, Miyata T, Mokuda S, Asahara H. Miyata K, et al. Methods Mol Biol. 2017;1668:3-13. doi: 10.1007/978-1-4939-7283-8_1. Methods Mol Biol. 2017. PMID: 28842898 Free PMC article. - Bisulfite methylation profiling of large genomes.
Reinders J, Paszkowski J. Reinders J, et al. Epigenomics. 2010 Apr;2(2):209-20. doi: 10.2217/epi.10.6. Epigenomics. 2010. PMID: 22121871 Review.
Cited by
- Detection of evolutionary conserved and accelerated genomic regions related to adaptation to thermal niches in Anolis lizards.
Sakamoto F, Kanamori S, Díaz LM, Cádiz A, Ishii Y, Yamaguchi K, Shigenobu S, Nakayama T, Makino T, Kawata M. Sakamoto F, et al. Ecol Evol. 2024 Mar 7;14(3):e11117. doi: 10.1002/ece3.11117. eCollection 2024 Mar. Ecol Evol. 2024. PMID: 38455144 Free PMC article. - Uncovering epigenetic and transcriptional regulation of growth in Douglas-fir: identification of differential methylation regions in mega-sized introns.
Vu GTH, Cao HX, Hofmann M, Steiner W, Gailing O. Vu GTH, et al. Plant Biotechnol J. 2024 Apr;22(4):863-875. doi: 10.1111/pbi.14229. Epub 2023 Nov 20. Plant Biotechnol J. 2024. PMID: 37984804 Free PMC article. - The Progression in Developing Genomic Resources for Crop Improvement.
Ruperao P, Rangan P, Shah T, Thakur V, Kalia S, Mayes S, Rathore A. Ruperao P, et al. Life (Basel). 2023 Jul 31;13(8):1668. doi: 10.3390/life13081668. Life (Basel). 2023. PMID: 37629524 Free PMC article. Review. - Introduction of a multiplex amplicon sequencing assay to quantify DNA methylation in target cytosine markers underlying four selected epigenetic clocks.
Pośpiech E, Pisarek A, Rudnicka J, Noroozi R, Boroń M, Masny A, Wysocka B, Migacz-Gruszka K, Lisman D, Pruszkowska-Przybylska P, Kobus M, Szargut M, Dowejko J, Stanisz K, Zacharczuk J, Zieliński P, Sitek A, Ossowski A, Spólnicka M, Branicki W. Pośpiech E, et al. Clin Epigenetics. 2023 Aug 10;15(1):128. doi: 10.1186/s13148-023-01545-2. Clin Epigenetics. 2023. PMID: 37563670 Free PMC article. - Methylated SEPT9 combined with AFP and PIVKA-II is effective for the detection of HCC in high-risk population.
Zheng K, Dai L, Zhao Y, Li L, Li W, Zhang X, Su Q, Wu R, Jiang Y, Chen Y, Ran J. Zheng K, et al. BMC Gastroenterol. 2023 Jul 31;23(1):260. doi: 10.1186/s12876-023-02900-6. BMC Gastroenterol. 2023. PMID: 37525116 Free PMC article.
References
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. - PubMed
- Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources