Emergence and evolution of multiply antibiotic-resistant Salmonella enterica serovar Paratyphi B D-tartrate-utilizing strains containing SGI1 - PubMed (original) (raw)
Emergence and evolution of multiply antibiotic-resistant Salmonella enterica serovar Paratyphi B D-tartrate-utilizing strains containing SGI1
Steven P Djordjevic et al. Antimicrob Agents Chemother. 2009 Jun.
Abstract
The first Australian isolate of Salmonella enterica serovar Paratyphi B D-tartrate-utilizing (dT(+)) that is resistant to ampicillin, chloramphenicol, florfenicol, streptomycin, spectinomycin, sulfonamides, and tetracycline (ApCmFlSmSpSuTc) and contains SGI1 was isolated from a patient with gastroenteritis in early 1995. This is the earliest reported isolation globally. The incidence of infections caused by these SGI1-containing multiply antibiotic-resistant S. enterica serovar Paratyphi B dT(+) strains increased during the next few years and occurred sporadically in all states of Australia. Several molecular criteria were used to show that the early isolates are very closely related to one another and to strains isolated during the following few years and in 2000 and 2003 from home aquariums and their owners. Early isolates from travelers returning from Indonesia shared the same features. Thus, they appear to represent a true clone arising from a single cell that acquired SGI1. Some minor differences in the resistance profiles and molecular profiles also were observed, indicating the ongoing evolution of the clone, and phage type differences were common, indicating that this is not a useful epidemiological marker over time. Three isolates from 1995, 1998, and 1999 contained a complete sul1 gene but were susceptible to sulfamethoxazole due to a point mutation that creates a premature termination codon. This SGI1 type was designated SGI1-R. The loss of resistance genes also was examined. When strains were grown for many generations in the absence of antibiotic selection, the loss of SGI1 was not detected. However, variants SGI1-C (resistance profile SmSpSu) and SGI1-B (resistant to ApSu), which had lost part of the integron, arose spontaneously, presumably via homologous recombination between duplications in the In104 complex integron.
Figures
FIG. 1.
Structure of the integron of SGI1 and deletion derivatives SGI1-B and SGI1-C. The In104 region of SGI1 serovar Typhimurium DT104 (from GenBank accession no. AF261825) (4) is drawn to scale. Different discrete segments, such as the gene cassettes, are represented by open boxes and lines of different thicknesses, and in the SGI1 line, 5′ and 3′ indicate the 5′-CS- and 3′-CS-derived regions, respectively. Vertical bars indicate the inverted repeats (IRi and IRt) of class 1 integrons bounding In104. The attI1 site is a tall open box, and gene cassettes (aadA2 and blaP1) are open boxes with a black bar at one end, indicating the attC sites (59-be). IS_6100_ and the central, non-integron-derived region are open boxes. Arrows indicate the position and orientation of genes and open reading frames. The SGI1 backbone adjacent to the integron is indicated by dashed lines.
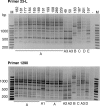
FIG. 2.
RAPD profiles of various serovar Paratyphi B dT+ strains. Whole-cell DNA was amplified with primer 23L (upper panel) or primer 1290 (lower panel), and the products were separated on a 1% agarose gel and stained with ethidium bromide. SRC numbers are above the gels, and designated types are shown below. Strains contain SGI1, except for SRC183, SRC187, SRC197, and SRC201, which were used as controls.
FIG. 3.
Alignment of the sequences at the boundaries of SGI elements with sequences from the chromosome of Salmonella serovar Paratyphi strains lacking SGI1. The sequences of the chromosome to the left and the right of SGI1 are separated by a short segment from SGI1, which is shown in italics. This region is dotted when SGI1 is not present. The remainder of the sequence internal to the SGI, which is identical in all sequences, is not shown. PtB+SGI1 represents sequences from SRC181 and SRC182 from 1995, SRC177 from 1997, SRC178 from 1998, SRC199 from 1999, SRC289 from 2000, and SRC49 from 2001; PtB represents sequences from four multidrug-resistant Salmonella serovar Paratyphi B dT+ strains lacking SGI1, SRC183, SRC187, SRC197, and SRC201, and PtB 272 is the sequence from a further multidrug-resistant Salmonella serovar Paratyphi B dT+ strain lacking SGI1, SRC272. Sequences from other SGI1-containing serovars were included for comparison. Kentucky represents the boundaries of SGI1-K from serovar Kentucky (GenBank accession no. AY463797), and Emek represents the boundaries of SGI2 from serovar Emek (GenBank accession no. AY963803). Derby W4 and W7 sequences, representing boundaries with SGI1-A and SGI1-C, respectively, are from reference . DT104 represents the equivalent regions of SGI1 from serovar Typhimurium DT104 (GenBank accession no. AF261825), and dotted lines indicate bases that are not present; the gap labeled retron phage delineates a segment that is missing from serovar Typhimurium. PtB SPB6 is from the genome sequence of strain SPB7 (GenBank accession no. CP000886). The initiation codon for the yidY gene and primers located in the chromosomal region and used to generate the PCR products sequenced are underlined. The recombination crossover region (shown in boldface), which includes the termination codon of the thdF gene (underlined), marks the boundaries between the SGI and the chromosome. Differences from the SGI1-K sequence are white on black.
Similar articles
- Isolation of Salmonella enterica subspecies enterica serovar Paratyphi B dT+, or Salmonella Java, from Indonesia and alteration of the d-tartrate fermentation phenotype by disrupting the ORF STM 3356.
Han KH, Choi SY, Lee JH, Lee H, Shin EH, Agtini MD, von Seidlein L, Ochiai RL, Clemens JD, Wain J, Hahn JS, Lee BK, Song M, Chun J, Kim DW. Han KH, et al. J Med Microbiol. 2006 Dec;55(Pt 12):1661-1665. doi: 10.1099/jmm.0.46792-0. J Med Microbiol. 2006. PMID: 17108269 - Multiple-antibiotic resistance in Salmonella enterica serotype Paratyphi B isolates collected in France between 2000 and 2003 is due mainly to strains harboring Salmonella genomic islands 1, 1-B, and 1-C.
Weill FX, Fabre L, Grandry B, Grimont PA, Casin I. Weill FX, et al. Antimicrob Agents Chemother. 2005 Jul;49(7):2793-801. doi: 10.1128/AAC.49.7.2793-2801.2005. Antimicrob Agents Chemother. 2005. PMID: 15980351 Free PMC article. - Antimicrobial resistance and virulence determinants in European Salmonella genomic island 1-positive Salmonella enterica isolates from different origins.
Beutlich J, Jahn S, Malorny B, Hauser E, Hühn S, Schroeter A, Rodicio MR, Appel B, Threlfall J, Mevius D, Helmuth R, Guerra B; Med-Vet-Net WP21 Project Group. Beutlich J, et al. Appl Environ Microbiol. 2011 Aug 15;77(16):5655-64. doi: 10.1128/AEM.00425-11. Epub 2011 Jun 24. Appl Environ Microbiol. 2011. PMID: 21705546 Free PMC article. - Salmonella genomic islands and antibiotic resistance in Salmonella enterica.
Hall RM. Hall RM. Future Microbiol. 2010 Oct;5(10):1525-38. doi: 10.2217/fmb.10.122. Future Microbiol. 2010. PMID: 21073312 Review. - The genetics of Salmonella genomic island 1.
Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A. Mulvey MR, et al. Microbes Infect. 2006 Jun;8(7):1915-22. doi: 10.1016/j.micinf.2005.12.028. Epub 2006 Mar 27. Microbes Infect. 2006. PMID: 16713724 Review.
Cited by
- Antimicrobial Resistance in Salmonella enterica Serovar Paratyphi B Variant Java in Poultry from Europe and Latin America.
Castellanos LR, van der Graaf-van Bloois L, Donado-Godoy P, Veldman K, Duarte F, Acuña MT, Jarquín C, Weill FX, Mevius DJ, Wagenaar JA, Hordijk J, Zomer AL. Castellanos LR, et al. Emerg Infect Dis. 2020 Jun;26(6):1164-1173. doi: 10.3201/eid2606.191121. Emerg Infect Dis. 2020. PMID: 32441616 Free PMC article. - Evaluation of the Safety and Protection Efficacy of spiC and nmpC or rfaL Deletion Mutants of Salmonella Enteritidis as Live Vaccine Candidates for Poultry Non-Typhoidal Salmonellosis.
Li Q, Zhu Y, Ren J, Qiao Z, Yin C, Xian H, Yuan Y, Geng S, Jiao X. Li Q, et al. Vaccines (Basel). 2019 Nov 30;7(4):202. doi: 10.3390/vaccines7040202. Vaccines (Basel). 2019. PMID: 31801257 Free PMC article. - Stability, entrapment and variant formation of Salmonella genomic island 1.
Kiss J, Nagy B, Olasz F. Kiss J, et al. PLoS One. 2012;7(2):e32497. doi: 10.1371/journal.pone.0032497. Epub 2012 Feb 23. PLoS One. 2012. PMID: 22384263 Free PMC article. - The master regulator of IncA/C plasmids is recognized by the Salmonella Genomic island SGI1 as a signal for excision and conjugal transfer.
Kiss J, Papp PP, Szabó M, Farkas T, Murányi G, Szakállas E, Olasz F. Kiss J, et al. Nucleic Acids Res. 2015 Oct 15;43(18):8735-45. doi: 10.1093/nar/gkv758. Epub 2015 Jul 24. Nucleic Acids Res. 2015. PMID: 26209134 Free PMC article. - Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota.
Djordjevic SP, Stokes HW, Roy Chowdhury P. Djordjevic SP, et al. Front Microbiol. 2013 Apr 30;4:86. doi: 10.3389/fmicb.2013.00086. eCollection 2013. Front Microbiol. 2013. PMID: 23641238 Free PMC article.
References
- Ahmed, A. M., K. Furuta, K. Shimomura, H. Kawamoto, and T. Shimamoto. 2005. Characterization of a multidrug-resistant isolate of Salmonella Paratyphi B from Japan. J. Antimicrob. Chemother. 56:250-251. - PubMed
- Ahmed, A. M., A. I. Hussein, and T. Shimamoto. 2007. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 59:184-190. - PubMed
- Akiba, M., K. Nakamura, D. Shinoda, N. Yoshii, H. Ito, I. Uchida, and M. Nakazawa. 2006. Detection and characterization of variant Salmonella genomic island 1s from Salmonella Derby isolates. Jpn. J. Infect. Dis. 59:341-345. - PubMed
- Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources