Human Splicing Finder: an online bioinformatics tool to predict splicing signals - PubMed (original) (raw)
Human Splicing Finder: an online bioinformatics tool to predict splicing signals
François-Olivier Desmet et al. Nucleic Acids Res. 2009 May.
Abstract
Thousands of mutations are identified yearly. Although many directly affect protein expression, an increasing proportion of mutations is now believed to influence mRNA splicing. They mostly affect existing splice sites, but synonymous, non-synonymous or nonsense mutations can also create or disrupt splice sites or auxiliary cis-splicing sequences. To facilitate the analysis of the different mutations, we designed Human Splicing Finder (HSF), a tool to predict the effects of mutations on splicing signals or to identify splicing motifs in any human sequence. It contains all available matrices for auxiliary sequence prediction as well as new ones for binding sites of the 9G8 and Tra2-beta Serine-Arginine proteins and the hnRNP A1 ribonucleoprotein. We also developed new Position Weight Matrices to assess the strength of 5' and 3' splice sites and branch points. We evaluated HSF efficiency using a set of 83 intronic and 35 exonic mutations known to result in splicing defects. We showed that the mutation effect was correctly predicted in almost all cases. HSF could thus represent a valuable resource for research, diagnostic and therapeutic (e.g. therapeutic exon skipping) purposes as well as for global studies, such as the GEN2PHEN European Project or the Human Variome Project.
Figures
Figure 1.
Branch point matrix. The size of each nucleotide is proportional to its weight in the position weight matrix. Nucleotides above the base line have positive values while nucleotides below have negative values.
Figure 2.
New position weight matrices of recognition motifs for proteins involved in splicing. (A) hnRNP A1; (B) Tra2-β and (C) 9G8.
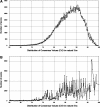
Figure 3.
Distribution of CVs for (A) 3′ and (B) 5′ natural splice sites (5′ss and 3′ss). Data extracted from the Ensembl dataset (release 44,
http://april2007.archive.ensembl.org
) (20) using the HSF algorithm.
Similar articles
- Splicing defects caused by exonic mutations in PKD1 as a new mechanism of pathogenesis in autosomal dominant polycystic kidney disease.
Claverie-Martin F, Gonzalez-Paredes FJ, Ramos-Trujillo E. Claverie-Martin F, et al. RNA Biol. 2015;12(4):369-74. doi: 10.1080/15476286.2015.1014291. RNA Biol. 2015. PMID: 25757501 Free PMC article. - Bioinformatics and mutations leading to exon skipping.
Desmet FO, Béroud C. Desmet FO, et al. Methods Mol Biol. 2012;867:17-35. doi: 10.1007/978-1-61779-767-5_2. Methods Mol Biol. 2012. PMID: 22454052 Review. - Skipping of an exon with a nonsense mutation in the DMD gene is induced by the conversion of a splicing enhancer to a splicing silencer.
Zhu Y, Deng H, Chen X, Li H, Yang C, Li S, Pan X, Tian S, Feng S, Tan X, Matsuo M, Zhang Z. Zhu Y, et al. Hum Genet. 2019 Jul;138(7):771-785. doi: 10.1007/s00439-019-02036-2. Epub 2019 Jun 5. Hum Genet. 2019. PMID: 31168774 - ESEfinder: A web resource to identify exonic splicing enhancers.
Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. Cartegni L, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3568-71. doi: 10.1093/nar/gkg616. Nucleic Acids Res. 2003. PMID: 12824367 Free PMC article. - Rules and tools to predict the splicing effects of exonic and intronic mutations.
Ohno K, Takeda JI, Masuda A. Ohno K, et al. Wiley Interdiscip Rev RNA. 2018 Jan;9(1). doi: 10.1002/wrna.1451. Epub 2017 Sep 26. Wiley Interdiscip Rev RNA. 2018. PMID: 28949076 Review.
Cited by
- U1snRNP-mediated suppression of polyadenylation in conjunction with the RNA structure controls poly (A) site selection in foamy viruses.
Schrom EM, Moschall R, Hartl MJ, Weitner H, Fecher D, Langemeier J, Bohne J, Wöhrl BM, Bodem J. Schrom EM, et al. Retrovirology. 2013 May 29;10:55. doi: 10.1186/1742-4690-10-55. Retrovirology. 2013. PMID: 23718736 Free PMC article. - Novel insights into the molecular pathogenesis of CYP4V2-associated Bietti's retinal dystrophy.
Astuti GD, Sun V, Bauwens M, Zobor D, Leroy BP, Omar A, Jurklies B, Lopez I, Ren H, Yazar V, Hamel C, Kellner U, Wissinger B, Kohl S, De Baere E, Collin RW, Koenekoop RK. Astuti GD, et al. Mol Genet Genomic Med. 2015 Jan;3(1):14-29. doi: 10.1002/mgg3.109. Epub 2014 Sep 15. Mol Genet Genomic Med. 2015. PMID: 25629076 Free PMC article. - Novel PRKAR1A mutation in Carney complex: a case report and literature review.
Zheng H, Kang H, Qiu Y, Xie L, Wu J, Lai P, Kang J. Zheng H, et al. Front Endocrinol (Lausanne). 2024 Jul 10;15:1384956. doi: 10.3389/fendo.2024.1384956. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39050568 Free PMC article. Review. - Antisense oligonucleotide-mediated exon skipping as a strategy to reduce proteolytic cleavage of ataxin-3.
Toonen LJ, Schmidt I, Luijsterburg MS, van Attikum H, van Roon-Mom WM. Toonen LJ, et al. Sci Rep. 2016 Oct 12;6:35200. doi: 10.1038/srep35200. Sci Rep. 2016. PMID: 27731380 Free PMC article. - Molecular Basis and Therapeutic Strategies to Rescue Factor IX Variants That Affect Splicing and Protein Function.
Tajnik M, Rogalska ME, Bussani E, Barbon E, Balestra D, Pinotti M, Pagani F. Tajnik M, et al. PLoS Genet. 2016 May 26;12(5):e1006082. doi: 10.1371/journal.pgen.1006082. eCollection 2016 May. PLoS Genet. 2016. PMID: 27227676 Free PMC article.
References
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. - PubMed
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. - PubMed
- Breitbart RE, Nguyen HT, Medford RM, Destree AT, Mahdavi V, Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985;41:67–82. - PubMed
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases