Structural basis for leucine-rich nuclear export signal recognition by CRM1 - PubMed (original) (raw)
. 2009 Apr 30;458(7242):1136-41.
doi: 10.1038/nature07975. Epub 2009 Apr 1.
Affiliations
- PMID: 19339969
- PMCID: PMC3437623
- DOI: 10.1038/nature07975
Structural basis for leucine-rich nuclear export signal recognition by CRM1
Xiuhua Dong et al. Nature. 2009.
Erratum in
- Nature. 2009 Sep 24;461(7263):550
Abstract
CRM1 (also known as XPO1 and exportin 1) mediates nuclear export of hundreds of proteins through the recognition of the leucine-rich nuclear export signal (LR-NES). Here we present the 2.9 A structure of CRM1 bound to snurportin 1 (SNUPN). Snurportin 1 binds CRM1 in a bipartite manner by means of an amino-terminal LR-NES and its nucleotide-binding domain. The LR-NES is a combined alpha-helical-extended structure that occupies a hydrophobic groove between two CRM1 outer helices. The LR-NES interface explains the consensus hydrophobic pattern, preference for intervening electronegative residues and inhibition by leptomycin B. The second nuclear export signal epitope is a basic surface on the snurportin 1 nucleotide-binding domain, which binds an acidic patch on CRM1 adjacent to the LR-NES site. Multipartite recognition of individually weak nuclear export signal epitopes may be common to CRM1 substrates, enhancing CRM1 binding beyond the generally low affinity LR-NES. Similar energetic construction is also used in multipartite nuclear localization signals to provide broad substrate specificity and rapid evolution in nuclear transport.
Figures
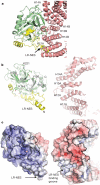
Figure 1. Overall structure of the CRM1–SNUPN complex
a, Orthogonal views of the CRM1–SNUPN complex. CRM1 is pink and the sIBB and NBD domains of SNUPN are yellow and green, respectively. HEAT repeats 2–20 of CRM1 are labelled H2–H20. b, HEAT repeat organization of CRM1. Most of H1 is disordered and not modelled in the structure. c, Structure of CRM1-bound SNUPN. The linker between sIBB and NBD of SNUPN is shown as a dashed yellow line and a grey arrow marks the location of the nucleotide-binding site of this nucleotide-free SNUPN.
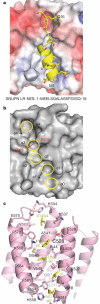
Figure 2. The LR-NES-binding site
a, The LR-NES interface showing the electrostatic surface potential of CRM1 and a ribbon diagram of the SNUPN LR-NES. The amino-acid sequence of the LR-NES of SNUPN is shown. b, Molecular surface of the LR-NES binding groove. Individual pockets that bind the five hydrophobic residues of the SNUPN LR-NES are outlined in yellow. Additional pockets are labelled and a dashed line is drawn between Lys 568 and Glu 529 at the narrowest point of the groove. c, Residues in the hydrophobic groove of CRM1 (pink) and hydrophobic residues of the SNUPN LR-NES (yellow).
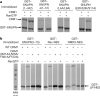
Figure 3. Hydrophobic residues are critical for LR-NES recognition
a, Mutations in the LR-NES of SNUPN that affected CRM1 binding. Deletion of the LR-NES (SNUPNΔ(1–12)) and mutation of residues Leu 4 and Leu 8 (SNUPN(L4A/L8A)) abolished interaction with CRM1. Mutation of electronegative residues of the LR-NES (SNUPN(E2K/E3K/S11A)) also decreased CRM1 binding. Bound proteins in the in vitro pull-down assays of immobilized GST–SNUPN proteins and CRM1 were visualized by SDS–PAGE and Coomassie staining. b, Mutations of hydrophobic residues on CRM1 helices H11A and H12A disrupt interaction with LR-NESs. Immobilized GST–NESs of SNUPN, HIV-1 Rev and NMD3 were incubated with either wild-type (WT) CRM1 or mutant CRM1(I521A/L525A/F561A/ F572A) in the presence and absence of RanGTP. Bound proteins were resolved with SDS–PAGE and Coomassie staining. MW stds, molecular weight standards.
Figure 4. Localization of the SNUPN and its LR-NES in yeast
a, The NLSGFP–GFP–SNUPN reporter is localized to the cytoplasm in the wild-type S. cerevisiae cells, but the NLS-GFP–GFP–SNUPN(L4A/L8A) mutant accumulates in the nucleus. Each panel displays the same GFP fluorescence scale. b, Location of NLS-GFP–GFP–SNUPN-NES in xpo1-1 temperature-sensitive (top) and wild-type (bottom) cells at the permissive (room temperature, RT) and non-permissive (37 °C) temperature. The reporter accumulates in the nucleus of xpo1-1 temperature-sensitive cells at the non-permissive temperature, but remains cytoplasmic in the wild-type cells.
Figure 5. Interactions of CRM1 with the SNUPN NES epitope II
a, Ribbon drawing of the interface between CRM1 (pink) and the NES epitope II of SNUPN, which is found on its NBD (green). The N-terminal sIBB domain of SNUPN is in yellow. No nucleotide is bound in the CRM1–SNUPN complex but the m3G-cap nucleotide from superimposed nucleotide-bound NBD (PDB accession 1XK5) is drawn in grey to show the location of the nucleotide-binding site. b, c, SNUPN and CRM1 in a are pried apart to show the interface (outlined with black lines) and electrostatic surface potential (scale of –12 kT e–1 to +12 kT e–1) of individual proteins at the interface.
Similar articles
- Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals.
Fung HY, Fu SC, Chook YM. Fung HY, et al. Elife. 2017 Mar 10;6:e23961. doi: 10.7554/eLife.23961. Elife. 2017. PMID: 28282025 Free PMC article. - Structural determinants of nuclear export signal orientation in binding to exportin CRM1.
Fung HY, Fu SC, Brautigam CA, Chook YM. Fung HY, et al. Elife. 2015 Sep 8;4:e10034. doi: 10.7554/eLife.10034. Elife. 2015. PMID: 26349033 Free PMC article. - Correlation of CRM1-NES affinity with nuclear export activity.
Fu SC, Fung HYJ, Cağatay T, Baumhardt J, Chook YM. Fu SC, et al. Mol Biol Cell. 2018 Aug 15;29(17):2037-2044. doi: 10.1091/mbc.E18-02-0096. Epub 2018 Jun 21. Mol Biol Cell. 2018. PMID: 29927350 Free PMC article. - Atomic basis of CRM1-cargo recognition, release and inhibition.
Fung HY, Chook YM. Fung HY, et al. Semin Cancer Biol. 2014 Aug;27:52-61. doi: 10.1016/j.semcancer.2014.03.002. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631835 Free PMC article. Review. - Inhibition of CRM1-dependent nuclear export sensitizes malignant cells to cytotoxic and targeted agents.
Turner JG, Dawson J, Cubitt CL, Baz R, Sullivan DM. Turner JG, et al. Semin Cancer Biol. 2014 Aug;27:62-73. doi: 10.1016/j.semcancer.2014.03.001. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631834 Free PMC article. Review.
Cited by
- Nuclear import of UBL-domain protein Mdy2 is required for heat-induced stress response in Saccharomyces cerevisiae.
Arhzaouy K, Ramezani-Rad M. Arhzaouy K, et al. PLoS One. 2012;7(12):e52956. doi: 10.1371/journal.pone.0052956. Epub 2012 Dec 28. PLoS One. 2012. PMID: 23285234 Free PMC article. - Nuclear transport proteins: structure, function, and disease relevance.
Yang Y, Guo L, Chen L, Gong B, Jia D, Sun Q. Yang Y, et al. Signal Transduct Target Ther. 2023 Nov 10;8(1):425. doi: 10.1038/s41392-023-01649-4. Signal Transduct Target Ther. 2023. PMID: 37945593 Free PMC article. Review. - Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A.
Aksu M, Trakhanov S, Görlich D. Aksu M, et al. Nat Commun. 2016 Jun 16;7:11952. doi: 10.1038/ncomms11952. Nat Commun. 2016. PMID: 27306458 Free PMC article. - NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, Ficner R, Sattler M, Görlich D. Güttler T, et al. Nat Struct Mol Biol. 2010 Nov;17(11):1367-76. doi: 10.1038/nsmb.1931. Epub 2010 Oct 24. Nat Struct Mol Biol. 2010. PMID: 20972448 - Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1.
Zhang ZC, Satterly N, Fontoura BM, Chook YM. Zhang ZC, et al. Mol Biol Cell. 2011 Dec;22(23):4657-68. doi: 10.1091/mbc.E11-03-0222. Epub 2011 Sep 30. Mol Biol Cell. 2011. PMID: 21965294 Free PMC article.
References
- Tran EJ, Bolger TA, Wente SR. SnapShot: nuclear transport. Cell. 2007;131:420. - PubMed
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. - PubMed
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. - PubMed
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 2001;13:310–319. - PubMed
- Dingwall C, Sharnick SV, Laskey RA. A polypeptide domain that specifies migration of nucleoplasmin into the nucleus. Cell. 1982;30:449–458. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM069909-03S1/GM/NIGMS NIH HHS/United States
- R01GM069909/GM/NIGMS NIH HHS/United States
- T32 GM008297/GM/NIGMS NIH HHS/United States
- 5-T32-GM008297/GM/NIGMS NIH HHS/United States
- R01 GM069909/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases