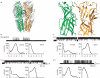Detection and trapping of intermediate states priming nicotinic receptor channel opening - PubMed (original) (raw)
Detection and trapping of intermediate states priming nicotinic receptor channel opening
Nuriya Mukhtasimova et al. Nature. 2009.
Abstract
In the course of synaptic transmission in the brain and periphery, acetylcholine receptors (AChRs) rapidly transduce a chemical signal into an electrical impulse. The speed of transduction is facilitated by rapid ACh association and dissociation, suggesting a binding site relatively non-selective for small cations. Selective transduction has been thought to originate from the ability of ACh, over that of other organic cations, to trigger the subsequent channel-opening step. However, transitions to and from the open state were shown to be similar for agonists with widely different efficacies. By studying mutant AChRs, we show here that the ultimate closed-to-open transition is agonist-independent and preceded by two primed closed states; the first primed state elicits brief openings, whereas the second elicits long-lived openings. Long-lived openings and the associated primed state are detected in the absence and presence of an agonist, and exhibit the same kinetic signatures under both conditions. By covalently locking the agonist-binding sites in the bound conformation, we find that each site initiates a priming step. Thus, a change in binding-site conformation primes the AChR for channel opening in a process that enables selective activation by ACh while maximizing the speed and efficiency of the biological response.
Figures
Fig. 1
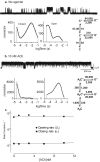
Agonist-independent channel gating. a, Spontaneous single channel currents through AChR containing Leu-to-Ser mutations at position 9′ of the second transmembrane domain (L9’S) of the β and δ subunits. Cell-attached configuration; membrane potential, -70 mV; bandwidth, 10 kHz; channel openings are upward deflections. Dwell time histograms are shown with probability density functions obtained by fitting the scheme, right, to the dwell times. Rate constants are from Table S1. b, Same as panel a but with 10 nM ACh. c, Plot of fitted channel opening and closing rate constants against ACh concentration (Table S1).
Fig. 2
Covalent priming of the AChR. a, Torpedo AChR (PDB code: 2bg9); α-subunit green, δ-subunit orange. Boxed region, magnified right, indicates Cys substitutions at positions α192 and δ121. b, upper trace: spontaneous currents through AChR containing L9’S mutations in the β and δ subunits and Cys substitutions at both ACh binding sites. Lower trace: Spontaneous currents from the patch above following application of H2O2. c, upper trace: spontaneous currents through AChR with L9’S mutations and Cys substitutions at both ACh binding sites treated with H2O2. Lower trace: Spontaneous currents from the patch above following application of DTT. In panels b and c, dwell time histograms are fitted by sums of exponentials. Results are summarized in Table S2.
Fig. 3
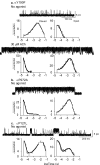
Mutating residues linking binding and pore domains increases or decreases priming. a, Upper trace: spontaneous single channel currents through AChR containing L9’S mutations in the β and δ subunits and αY190F. Lower trace: ACh-evoked single channel currents from a different patch containing the same mutant AChR. b, Spontaneous currents through AChR containing L9’S mutations in the β and δ subunits and αP272A. c, Spontaneous currents through AChR containing L9’S mutations in the β and δ subunits and εP121L. Dwell time histograms are shown with fitted probability density functions (Table S1).
Fig. 4
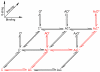
Primed model of AChR activation. Agonist binding, priming and channel gating steps are indicated (inset). C, C’ and C” symbolize closed states, while O’ and O” symbolize open states. For the wild type AChR in the absence of ACh, the C’ and C” states are negligible, indicating the first step in the activation process generates AC, from which there are three possible paths toward A2C”. Fitting the path, bind-bind-prime-prime, did not give well defined rate constants, possibly due to an inability to distinguish interconnected A2C’ and A2C” states. Fitting the path, bind-prime-prime-bind, also did not give well defined rate constants, possibly because the second binding step would be to a primed site presumed to have reduced accessibility to small molecules. The remaining path in red was fitted to agonist-dependent dwell times from the wild type AChR, yielding the rate constants in Table S4.
Similar articles
- Differential pharmacological activity of JN403 between α7 and muscle nicotinic acetylcholine receptors.
Arias HR, De Rosa MJ, Bergé I, Feuerbach D, Bouzat C. Arias HR, et al. Biochemistry. 2013 Nov 26;52(47):8480-8. doi: 10.1021/bi4012572. Epub 2013 Nov 11. Biochemistry. 2013. PMID: 24164482 - Site specificity of agonist-induced opening and desensitization of the Torpedo californica nicotinic acetylcholine receptor.
Andreeva IE, Nirthanan S, Cohen JB, Pedersen SE. Andreeva IE, et al. Biochemistry. 2006 Jan 10;45(1):195-204. doi: 10.1021/bi0516024. Biochemistry. 2006. PMID: 16388595 - Activation of endplate nicotinic acetylcholine receptors by agonists.
Auerbach A. Auerbach A. Biochem Pharmacol. 2015 Oct 15;97(4):601-608. doi: 10.1016/j.bcp.2015.06.024. Epub 2015 Jul 20. Biochem Pharmacol. 2015. PMID: 26206191 Free PMC article. Review. - Topology of ligand binding sites on the nicotinic acetylcholine receptor.
Arias HR. Arias HR. Brain Res Brain Res Rev. 1997 Oct;25(2):133-91. doi: 10.1016/s0165-0173(97)00020-9. Brain Res Brain Res Rev. 1997. PMID: 9403137 Review.
Cited by
- Zolpidem and eszopiclone prime α1β2γ2 GABAA receptors for longer duration of activity.
Dixon CL, Harrison NL, Lynch JW, Keramidas A. Dixon CL, et al. Br J Pharmacol. 2015 Jul;172(14):3522-36. doi: 10.1111/bph.13142. Epub 2015 May 11. Br J Pharmacol. 2015. PMID: 25817320 Free PMC article. - Partial agonism of taurine at gamma-containing native and recombinant GABAA receptors.
Kletke O, Gisselmann G, May A, Hatt H, Sergeeva OA. Kletke O, et al. PLoS One. 2013 Apr 30;8(4):e61733. doi: 10.1371/journal.pone.0061733. Print 2013. PLoS One. 2013. PMID: 23637894 Free PMC article. - Nicotinic receptor transduction zone: invariant arginine couples to multiple electron-rich residues.
Mukhtasimova N, Sine SM. Mukhtasimova N, et al. Biophys J. 2013 Jan 22;104(2):355-67. doi: 10.1016/j.bpj.2012.12.013. Biophys J. 2013. PMID: 23442857 Free PMC article. - Bayesian Statistical Inference in Ion-Channel Models with Exact Missed Event Correction.
Epstein M, Calderhead B, Girolami MA, Sivilotti LG. Epstein M, et al. Biophys J. 2016 Jul 26;111(2):333-348. doi: 10.1016/j.bpj.2016.04.053. Biophys J. 2016. PMID: 27463136 Free PMC article. - Mechanisms of inhibition and activation of extrasynaptic αβ GABAA receptors.
Kasaragod VB, Mortensen M, Hardwick SW, Wahid AA, Dorovykh V, Chirgadze DY, Smart TG, Miller PS. Kasaragod VB, et al. Nature. 2022 Feb;602(7897):529-533. doi: 10.1038/s41586-022-04402-z. Epub 2022 Feb 9. Nature. 2022. PMID: 35140402 Free PMC article.
References
Methods References
- Lee BS, Gunn RB, Kopito RR. Functional differences among nonerythroid anion exchangers expressed in a transfected human cell line. J. Biol. Chem. 1991;266:11448–11454. - PubMed
- Ohno K, Wang H-L, Milone M, Bren N, Brengman J, Nakano S, Quiram P, Pruitt J, Sine SM, Engel AG. Congential myasthenic syndrome caused by decreased agonist binding affinity due to a mutation in the acetylcholine receptor ε subunit. Neuron. 1996;17:157–170. - PubMed
- Colquhoun D, Sigworth F. Fitting and statistical analysis of single channel records. In: Sakmann B, Neher E, editors. Single Channel Recording. Plenum Publishing Corp.; New York: 1983. pp. 191–264.
References
- Del Castillo J, Katz B. Interaction at end-plate receptors between different choline derivatives. Proc. R. Soc. Lond. B. 1957;146:369–381. - PubMed
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Archiv. 1981;391:85–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS031744/NS/NINDS NIH HHS/United States
- R01 NS031744-18/NS/NINDS NIH HHS/United States
- R37 NS031744/NS/NINDS NIH HHS/United States
- NS031744/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources