Structural basis for assembly and disassembly of the CRM1 nuclear export complex - PubMed (original) (raw)
Structural basis for assembly and disassembly of the CRM1 nuclear export complex
Xiuhua Dong et al. Nat Struct Mol Biol. 2009 May.
Abstract
CRM1 (or exportin 1, Xpo1) transports proteins out of the cell nucleus through the nuclear pore complex. In the cytoplasm, GTP hydrolysis and consequent dissociation of Ran from CRM1 releases low-affinity substrates, while additional factors facilitate release of high-affinity substrates. Here we provide a model for human CRM1 export complex assembly and disassembly through structural and biochemical analyses of CRM1 bound to the substrate snurportin 1 (SNUPN, also called snuportin 1).
Figures
Figure 1
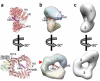
Comparison of unliganded and SNUPN-bound states of CRM1. (a) Ribbon diagram of the CRM1–SNUPN complex (PDB 3GB8). CRM1 is shown in pink and its C-extension in magenta, and several of the 20 HEAT repeats (H1–H20) are labeled. The sIBB and NBD domains of SNUPN are yellow and green, respectively. (b,c) Orthogonal views of the 22-Å-resolution density map calculated from coordinates of SNUPN-bound CRM1 (b) and the EM map of unliganded CRM1 (c). Ribbon diagram of SNUPN-bound CRM1 (blue-red color ramp from N to C terminus) is also shown in b, and a red triangle marks the LR-NES–binding site. N-terminal, central and C-terminal portions of CRM1 in the EM map are numbered in c.
Figure 2
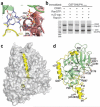
Positive cooperativity of substrate and Ran binding to CRM1. (a) The CRM1–SNUPN binary intermediate can accommodate RanGTP. Repeats H2–H7 of SNUPN-bound CRM1 were superimposed with H2–H7 of the CSE1–Kap60– RanGTP complex (PDB 1WA5). CRM1 and SNUPN are colored as in Figure 1 and RanGTP is in blue. CSE1 and Kap60 are omitted for clarity. (b) The electrostatic surface potential of SNUPN-bound CRM1 is shown with RanGTP from the superimposed CSE1–Kap60–Ran complex. The basic patch of Ran (residues 129–142) is in purple, the H9 loop sequence is shown and a dashed line indicates the disordered portion of this loop. (c) Pulldown assays with immobilized GST-NES of HIV1-REV and CRM1 proteins in the presence or absence of RanGTP and/or leptomycin B (LMB) were visualized by SDS-PAGE and Coomassie blue staining.
Figure 3
Comparison of CRM1-, Kap-β1- and m3G-cap-nucleotide-bound SNUPN proteins. (a) The m3G-cap binding site in the CRM1–SNUPN complex. The m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD is shown as sticks (PDB 1XK5). CRM1 is shown as a pink ribbon and the sIBB and NBD of SNUPN as yellow and green ribbons, respectively. (b) Pulldown assays using immobilized GSTSNUPN and CRM1 were performed in the presence of RanGTP, m3G-cap nucleotide or Kap-β1. Bound proteins were separated by SDS-PAGE and visualized with Coomassie blue staining. (c) The Kap-β1–sIBB complex (PDB 2P8Q) is shown with the molecular surface of Kap-β1 in gray and the sIBB as a yellow ribbon. (d) Ribbon diagram of CRM1-bound SNUPN, showing that most of the sIBB (yellow) interacts with its NBD (green). m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD is shown as sticks. The structure is drawn with residues 40–52 of the sIBB in the same orientation as in c.
Comment in
- Nuclear transport comes full circle.
Debler EW, Blobel G, Hoelz A. Debler EW, et al. Nat Struct Mol Biol. 2009 May;16(5):457-9. doi: 10.1038/nsmb0509-457. Nat Struct Mol Biol. 2009. PMID: 19421156 No abstract available.
Similar articles
- Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Dong X, Biswas A, Süel KE, Jackson LK, Martinez R, Gu H, Chook YM. Dong X, et al. Nature. 2009 Apr 30;458(7242):1136-41. doi: 10.1038/nature07975. Epub 2009 Apr 1. Nature. 2009. PMID: 19339969 Free PMC article. - Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP.
Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. Monecke T, et al. Science. 2009 May 22;324(5930):1087-91. doi: 10.1126/science.1173388. Epub 2009 Apr 23. Science. 2009. PMID: 19389996 - Nuclear transport comes full circle.
Debler EW, Blobel G, Hoelz A. Debler EW, et al. Nat Struct Mol Biol. 2009 May;16(5):457-9. doi: 10.1038/nsmb0509-457. Nat Struct Mol Biol. 2009. PMID: 19421156 No abstract available. - Atomic basis of CRM1-cargo recognition, release and inhibition.
Fung HY, Chook YM. Fung HY, et al. Semin Cancer Biol. 2014 Aug;27:52-61. doi: 10.1016/j.semcancer.2014.03.002. Epub 2014 Mar 12. Semin Cancer Biol. 2014. PMID: 24631835 Free PMC article. Review. - [Structural basis for assembly and disassembly of the CRM1 nuclear export complex and its application to drug development].
Koyama M, Matsuura Y. Koyama M, et al. Seikagaku. 2015 Feb;87(1):41-8. Seikagaku. 2015. PMID: 26571554 Review. Japanese. No abstract available.
Cited by
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA.
Gallmetzer A, Silvestrini L, Schinko T, Gesslbauer B, Hortschansky P, Dattenböck C, Muro-Pastor MI, Kungl A, Brakhage AA, Scazzocchio C, Strauss J. Gallmetzer A, et al. PLoS Genet. 2015 Jul 1;11(7):e1005297. doi: 10.1371/journal.pgen.1005297. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26132230 Free PMC article. - Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein.
Ishizawa J, Kojima K, Hail N Jr, Tabe Y, Andreeff M. Ishizawa J, et al. Pharmacol Ther. 2015 Sep;153:25-35. doi: 10.1016/j.pharmthera.2015.06.001. Epub 2015 Jun 3. Pharmacol Ther. 2015. PMID: 26048327 Free PMC article. Review. - A cellular reporter to evaluate CRM1 nuclear export activity: functional analysis of the cancer-related mutant E571K.
García-Santisteban I, Arregi I, Alonso-Mariño M, Urbaneja MA, Garcia-Vallejo JJ, Bañuelos S, Rodríguez JA. García-Santisteban I, et al. Cell Mol Life Sci. 2016 Dec;73(24):4685-4699. doi: 10.1007/s00018-016-2292-0. Epub 2016 Jun 16. Cell Mol Life Sci. 2016. PMID: 27312238 Free PMC article. - A structurally plastic ribonucleoprotein complex mediates post-transcriptional gene regulation in HIV-1.
Fernandes JD, Booth DS, Frankel AD. Fernandes JD, et al. Wiley Interdiscip Rev RNA. 2016 Jul;7(4):470-86. doi: 10.1002/wrna.1342. Epub 2016 Mar 1. Wiley Interdiscip Rev RNA. 2016. PMID: 26929078 Free PMC article. Review. - Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Sun Q, Carrasco YP, Hu Y, Guo X, Mirzaei H, Macmillan J, Chook YM. Sun Q, et al. Proc Natl Acad Sci U S A. 2013 Jan 22;110(4):1303-8. doi: 10.1073/pnas.1217203110. Epub 2013 Jan 7. Proc Natl Acad Sci U S A. 2013. PMID: 23297231 Free PMC article.
References
- Gorlich D, Kutay U. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. - PubMed
- Stade K, Ford CS, Guthrie C, Weis K. Cell. 1997;90:1041–1050. - PubMed
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. - PubMed
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M. Nature. 1997;390:308–311. - PubMed
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. Cell. 1997;90:1051–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM069909-03S1/GM/NIGMS NIH HHS/United States
- R01GM069909/GM/NIGMS NIH HHS/United States
- T32 GM008297/GM/NIGMS NIH HHS/United States
- 5-T32-GM008297/GM/NIGMS NIH HHS/United States
- R01 GM069909/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous