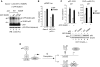Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding - PubMed (original) (raw)
Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding
Shwu-Yuan Wu et al. EMBO J. 2009.
Abstract
Covalent modification by small ubiquitin-related modifiers (SUMO) regulates p53 transcription activity through an undefined mechanism. Using reconstituted sumoylation components, we purified SUMO-1-conjugated p53 (Su-p53) to near homogeneity. Su-p53 exists in solution as a tetramer and interacts with p300 histone acetyltransferase as efficiently as the unmodified protein. Nevertheless, it fails to activate p53-dependent chromatin transcription because of its inability to bind DNA. With sequential modification assays, we found that sumoylation of p53 at K386 blocks subsequent acetylation by p300, whereas p300-acetylated p53 remains permissive for ensuing sumoylation at K386 and alleviates sumoylation-inhibited DNA binding. While preventing the free form of p53 from accessing its cognate sites, sumoylation fails to disengage prebound p53 from DNA. The sumoylation-deficient K386R protein, when expressed in p53-null cells, exhibits higher transcription activity and binds better to the endogenous p21 gene compared with the wild-type protein. These studies unravel a molecular mechanism underlying sumoylation-regulated p53 function and further uncover a new role of acetylation in antagonizing the inhibitory effect of sumoylation on p53 binding to DNA.
Figures
Figure 1
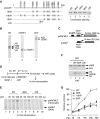
Acetylation of chromatin but not p53 is directly linked to p53-dependent transcription. (A) Coomassie blue staining of purified p53, 8KR, 10KR, 6KR and Δ30 proteins and their schematic drawing. Numbers indicate the positions of specific amino acid residues and the boundaries of each protein. (B) Coomassie blue staining of purified HeLa core histones (CH), hNAP-1 and ACF proteins. (C) G-less cassette templates used for in vitro transcription assays. (D) Scheme of chromatin assembly for histone acetyltransferase (HAT) assay and in vitro transcription. (E) Wild-type (WT) p53 and 8KR, but not Δ30, efficiently activate transcription from pWAFMLT chromatin. In vitro transcription was performed with pWAFMLT chromatin and the internal control pΔMLP DNA template as described in Materials and methods. (F) Wild-type p53 and 8KR, but not Δ30, support p300-mediated acetylation of pWAFMLT chromatin. In vitro HAT assay was performed with p300 and pWAFMLT chromatin, in the absence (−) or presence of different p53 proteins as indicated. (G) Dose-dependent activation of pWAFMLT chromatin by wild-type p53 and acetylation-defective mutants. Error bars indicate the standard deviation from an average of three independent experiments.
Figure 2
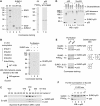
Purified SUMO-1-conjugated p53 exists in solution as a tetramer with selective C-termini accessible for sumoylation. (A) Coomassie blue staining of purified proteins used for in vitro sumoylation reactions. All recombinant human proteins contain an N-terminal hexahistidine tag. Purification of these hexahistidine-tagged proteins was described in Materials and methods. Molecular size markers (in kDa) are indicated on the left. (B) Sumoylated p53 (Su-p53) contains both SUMO-1-conjugated and unmodified subunits. Purification of Su-p53 was conducted as outlined (left), following a large-scale sumoylation reaction performed with (+) or without (−) the SUMO mix containing either FLAG (f:)- or hexahistidine (6His)-tagged sumoylation components. Purified Su-p53, along with the samples recovered from the mock purification, was visualized by Coomassie blue staining. (C) Su-p53 and unmodified p53 each elutes as a tetramer as determined by Superose 6 size-exclusion column chromatography. The void volume and predetermined protein size markers (in kDa) are indicated above the fraction numbers. (D) Tetrameric p53 and Su-p53 detected by glutaraldehyde crosslinking. An increasing concentration (%) of glutaraldehyde was used for crosslinking homomeric p53 subunits. Products were analysed by Western blotting with anti-p53 full-length antibodies. (E) Purification of Su-p53 with different extent of SUMO-modified subunits. Constant amounts of p53, E1 and E2 enzymes in a standard 1 × reaction were incubated with variable quantities of SUMO-1 and PIASxβ as indicated. The extent of p53 sumoylation, before and after purification (with schematic drawing on the right), was visualized after Coomassie blue staining. (F) Further sumoylation can occur on purified Su-p53. Constant amounts (1 × ) of Su-p53, SUMO-1 and PIASxβ were incubated with an increasing concentration (1–10 × ) of E1 and E2 as indicated. Western blotting was performed with anti-p53 antibodies.
Figure 3
Sumoylation inhibits p53-dependent transcription by preventing p53 from binding to DNA/chromatin, correlating with reduced p300-mediated acetylation on p53 and chromatin. (A) Quantification of protein amounts between p53 and Su-p53 by Western blotting. (B) Su-p53 fails to activate p53-dependent transcription from pWAFMLT chromatin. In vitro transcription was performed as described in Materials and methods using different amounts of purified p53 or Su-p53 as indicated. Relative transcription (%) is defined as the signal intensity from pWAFMLT chromatin relative to that of lane 5 (set at 100), after normalization of the transcription signal with that of the pΔMLP internal control. (C) Su-p53 could not be acetylated by p300 and could not efficiently support p300-mediated acetylation on pWAFMLT chromatin. In vitro HAT assay was performed with p300 and pWAFMLT chromatin, in the absence (−) or presence of different amounts of p53 or Su-p53 as indicated and described in Materials and methods. (D) Sumoylation does not affect p53 association with p300. Solution interaction was performed as outlined by incubating p53 or Su-p53 with p300 or buffer alone (−). Asterisk indicates the immunoglobulin heavy chain from anti-p300 antibodies that serve as a good internal control for normalization among different immunoprecipitated (IP) samples. (E) Su-p53 is incapable of binding to pWAFMLT chromatin. In vitro chromatin immunoprecipitation (ChIP) was conducted by incubating p53 or Su-p53 with p300 and pWAFMLT chromatin, followed by formaldehyde crosslinking and micrococcal nuclease digestion before further processing for ChIP analysis as described in Materials and methods. (F) Su-p53 is unable to bind DNA containing a p53-binding site. Electrophoretic mobility shift assay (EMSA) was carried out with a 32P-labelled DNA fragment containing the p53-binding site derived from pWAFMLT (i.e. the distal p53-binding site originally from the human p21 gene). (G) Su-p53 cannot bind to a p53-binding site-containing Hdm2 DNA fragment as determined by EMSA. (H) SENP1 unmasks the DNA-binding activity of Su-p53 after desumoylation. Purified Su-p53 was incubated with wild-type (WT) or a catalytic mutant (R630L/K631M; mt) of SENP1 and then analysed by Western blotting and EMSA using the same pWAFMLT-derived fragment as in (F). (I) Coomassie blue staining of purified WT and mt FLAG-tagged SENP1 used in (H).
Figure 4
Acetylation overcomes sumoylation-mediated inhibition of sequence recognition by p53. (A) Sequence-specific binding of p53 on immobilized DNA template. Binding of p53 to immobilized pWAFMLT DNA, with or without _Xba_I digestion, was monitored by Western blotting following the outlined protocol. (B) Coomassie blue staining of purified FLAG-tagged wild-type (WT) p53 and the 5KQ mutant. (C) Sumoylation blocks wild-type p53 but not 5KQ binding to immobilized pWAFMLT DNA. DNA-binding assays were performed as outlined by incubating wild-type p53 or the acetylation-mimic 5KQ mutant with immobilized pWAFMLT DNA (+) or beads alone (−), with (+) or without (−) prior sumoylation as indicated. (D) Acetylation does not prevent subsequent sumoylation of p53 and allows doubly modified p53 binding to DNA. Sequential acetylation and sumoylation reactions were carried out as outlined with the addition of desulfo-CoA in between to terminate the acetylation reaction before the sumoylation reaction. Acetylated p53 was monitored by incorporated 3H on the acetyl group. (E) DNA binding by acetylated Su-p53 containing different SUMO-modified subunits. Purified Su-p53 in 2:2 or 1:3 stoichiometry, as prepared in Figure 2E, with a minor population also acetylated by p300, was incubated in the presence (+) or absence (−) of pWAFMLT DNA. Western blotting performed with anti-p53 full-length (FL) or anti-acetylated K373/382 antibodies was used to monitor the DNA-binding activity of Su-p53 without acetylation (upper panel) or with p300-mediated acetylation at K373/382 (lower panel). (F) SENP1 treatment restores the DNA-binding activity of Su-p53 with 1:3 stoichiometry. Purified Su-p53 (1:3) was incubated in the presence (+) or absence (−) of SENP1 and then analysed by Western blotting and EMSA using the pWAFMLT-derived fragment.
Figure 5
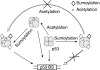
Model for the crosstalk between sumoylation and acetylation regulating p53 tetramer binding to its target sequence. The subunits modified by acetylation (asterisk) and/or sumoylation (diamond) are arbitrarily assigned to simply reflect the modified state of the C-termini with distinct conformations within p53 tetramers. The p53-binding site is abbreviated as p53 BS.
Figure 6
Sumoylation of p53 in the DNA-bound state occurs at K386 and fails to dissociate p53 from its target sequence. (A) Sumoylation takes place on DNA-bound p53 without leading to its dissociation from the immobilized pWAFMLT template. Sumoylation reactions were performed with wild-type SUMO-1 or the GA mutant on p53- or 5KQ-bound pWAFMLT DNA as outlined. Protein-bound beads (B) and the unbound supernatant (UB) were analysed by Western blotting with anti-p53 full-length antibodies. (B) Sumoylation of DNA-bound p53 occurs at the same K386 residue. Sumoylation of wild-type p53 or the K292R or K386R mutant prebound to immobilized pWAFMLT DNA was performed and analysed as outlined. (C) Enhanced sumoylation on DNA-bound p53 fails to disengage p53 from DNA. DNA-bound p53 was subjected to different extent of sumoylation reactions with 2 times (2 × ) or 5 times (5 × ) more of sumoylation components. Binding assay was conducted as described in (A). (D) Recruitment of mSin3A but not p300 is enhanced by sumoylated p53 bound to DNA. HeLa nuclear extract was incubated with DNA-bound p53, with (+) or without (−) prior sumoylation reactions performed with wild-type (WT) or the GA mutant of SUMO-1. Proteins associated with DNA-bound p53 or sumoylated p53 were then analysed by Western blotting with anti-p53, p300 or mSin3A antibodies.
Figure 7
Sumoylation-deficient p53 exhibits higher transcription activity and binds better to the endogenous p21 gene than the wild-type protein. (A) Detection of sumoylated p53 in p53-null Saos-2 cells by exogenous expression of p53 or the K386R mutant, with or without coexpressed GFP-tagged wild-type (WT) SUMO-1 (GFP-SUMO) or the diglycine-deleted ΔGG mutant. (B) Sumoylation-defective K386R exhibits higher transactivating activity compared with that of wild-type p53 in enhancing human p21 promoter-driven reporter (pWWP-Luc) activity in p53-null Saos-2 and HCT116 cells. (C) K386R activates endogenous p53-targeted p21 gene expression more efficiently than wild-type p53, correlating with enhanced K386 association with the p21 gene. RNA, chromatin, and cell lysates were harvested from HCT116 p53−/− cells exogenously expressing wild-type p53 or K386R, with (+) or without (−) coexpressed GFP-SUMO-1, and analysed by quantitative RT–PCR (left panel), ChIP (right panel) and Western blotting (bottom panel), respectively. (D) Model for sumoylation-regulated p53 target gene transcription. The transcription preinitiation complex (PIC) assembled on a p53-regulated promoter (left) could be dissociated from (upper right) or remained bound to (lower right) the promoter depending on sumoylation on either free or DNA-bound p53. A poised PIC, after sumoylation on DNA-bound p53 tetramers, is likely in an inactive state.
Similar articles
- SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment.
Mohan RD, Rao A, Gagliardi J, Tini M. Mohan RD, et al. Mol Cell Biol. 2007 Jan;27(1):229-43. doi: 10.1128/MCB.00323-06. Epub 2006 Oct 23. Mol Cell Biol. 2007. PMID: 17060459 Free PMC article. - Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases.
Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Barlev NA, et al. Mol Cell. 2001 Dec;8(6):1243-54. doi: 10.1016/s1097-2765(01)00414-2. Mol Cell. 2001. PMID: 11779500 - p53 sumoylation: mechanistic insights from reconstitution studies.
Wu SY, Chiang CM. Wu SY, et al. Epigenetics. 2009 Oct 1;4(7):445-51. doi: 10.4161/epi.4.7.10030. Epub 2009 Oct 9. Epigenetics. 2009. PMID: 19838051 Free PMC article. - Regulation of transcription factors by sumoylation.
Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS. Rosonina E, et al. Transcription. 2017 Aug 8;8(4):220-231. doi: 10.1080/21541264.2017.1311829. Epub 2017 Apr 5. Transcription. 2017. PMID: 28379052 Free PMC article. Review. - SUMO and Chromatin Remodeling.
Wotton D, Pemberton LF, Merrill-Schools J. Wotton D, et al. Adv Exp Med Biol. 2017;963:35-50. doi: 10.1007/978-3-319-50044-7_3. Adv Exp Med Biol. 2017. PMID: 28197905 Review.
Cited by
- MEL-18 loss mediates estrogen receptor-α downregulation and hormone independence.
Lee JY, Won HY, Park JH, Kim HY, Choi HJ, Shin DH, Kang JH, Woo JK, Oh SH, Son T, Choi JW, Kim S, Kim HY, Yi K, Jang KS, Oh YH, Kong G. Lee JY, et al. J Clin Invest. 2015 May;125(5):1801-14. doi: 10.1172/JCI73743. Epub 2015 Mar 30. J Clin Invest. 2015. PMID: 25822021 Free PMC article. - SUMO-mediated regulation of DNA damage repair and responses.
Sarangi P, Zhao X. Sarangi P, et al. Trends Biochem Sci. 2015 Apr;40(4):233-42. doi: 10.1016/j.tibs.2015.02.006. Epub 2015 Mar 13. Trends Biochem Sci. 2015. PMID: 25778614 Free PMC article. Review. - The RAX/PACT-PKR stress response pathway promotes p53 sumoylation and activation, leading to G₁ arrest.
Bennett RL, Pan Y, Christian J, Hui T, May WS Jr. Bennett RL, et al. Cell Cycle. 2012 Jan 15;11(2):407-17. doi: 10.4161/cc.11.2.18999. Epub 2012 Jan 15. Cell Cycle. 2012. PMID: 22214662 Free PMC article. - Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5.
Sun D, Xu P, He B. Sun D, et al. J Virol. 2011 Oct;85(19):10261-8. doi: 10.1128/JVI.00389-11. Epub 2011 Jul 27. J Virol. 2011. PMID: 21795356 Free PMC article. - Heat shock transcription factor 1 is SUMOylated in the activated trimeric state.
Kmiecik SW, Drzewicka K, Melchior F, Mayer MP. Kmiecik SW, et al. J Biol Chem. 2021 Jan-Jun;296:100324. doi: 10.1016/j.jbc.2021.100324. Epub 2021 Jan 23. J Biol Chem. 2021. PMID: 33493517 Free PMC article.
References
- Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772 - PubMed
- Baba D, Maita N, Jee J-G, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M (2006) Crystal structure of SUMO-3-modified thymine-DNA glycosylase. J Mol Biol 359: 137–147 - PubMed
- Carter S, Bischof O, Dejean A, Vousden KH (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nature Cell Biol 9: 428–435 - PubMed
- Čes̆ková P, Chichger H, Wallace M, Vojtesek B, Hupp TR (2006) On the mechanism of sequence-specific DNA-dependent acetylation of p53: the acetylation motif is exposed upon DNA binding. J Mol Biol 357: 442–456 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous