Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells - PubMed (original) (raw)
. 2009 Apr 3;4(4):359-69.
doi: 10.1016/j.stem.2009.03.001.
Linlin Wang, Brigham H Mecham, Lanlan Shen, Angelique M Nelson, Merav Bar, Deepak A Lamba, Derek S Dauphin, Brian Buckingham, Bardia Askari, Raymond Lim, Muneesh Tewari, Stanley M Gartler, Jean-Pierre Issa, Paul Pavlidis, Zhijun Duan, C Anthony Blau
Affiliations
- PMID: 19341625
- PMCID: PMC2719860
- DOI: 10.1016/j.stem.2009.03.001
Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells
Carol B Ware et al. Cell Stem Cell. 2009.
Abstract
Recent evidence indicates that mouse and human embryonic stem cells (ESCs) are fixed at different developmental stages, with the former positioned earlier. We show that a narrow concentration of the naturally occurring short-chain fatty acid, sodium butyrate, supports the extensive self-renewal of mouse and human ESCs, while promoting their convergence toward an intermediate stem cell state. In response to butyrate, human ESCs regress to an earlier developmental stage characterized by a gene expression profile resembling that of mouse ESCs, preventing precocious Xist expression while retaining the ability to form complex teratomas in vivo. Other histone deacetylase inhibitors (HDACi) also support human ESC self-renewal. Our results indicate that HDACi can promote ESC self-renewal across species, and demonstrate that ESCs can toggle between alternative states in response to environmental factors.
Figures
Figure 1
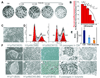
Butyrate supports the self-renewal of H1 cells. A. H1 cells acclimated to growth in CM (which contains 2ng/ml FGF2-see methods) were cultured on Matrigel-coated 35 mm dishes for 4 days in conditioned medium (CM) or in hESM (which lacks conditioning or FGF2) at the indicated concentrations of butyrate. Undifferentiated colonies were scored based on alkaline phosphatase staining. B. Quantitative results from a repeat of the experiment shown in panel a), performed in triplicate. Black line indicates numbers of colonies per dish. * = p<0.05, ** = p<0.01. Red bars indicate percentages of alkaline phosphatase-positive colonies. Error bars denote standard errors of the mean. C. Appearance of differentiated cells that appear transiently after switching from CM to butyrate for two passages. D. Cell cycle profiles of H1 cells cultured in CM (left) versus butyrate (right). Numbers indicate percentages of cells in G1/S/G2. Note increases in percentages of cells in S and G2 in the butyrate cultures. These findings are representative of 2 independent experiments. E. Bromodeoxyuridine (BrdU) incorporation in H1 cells cultured in CM, trichostatin A (TSA) (10 nM) (*P<0.05; CM vs. TSA) and butyrate (0.2mM) (***P<0.001; CM vs. butyrate) F. Morphology of H1 cells cultured in CM (1–5) or butyrate (6–10). 1,6: on feeders; 2–5 and 7–10: on MatrigelTM; 3, 8: Oil red O stained; 4,9: Pou5F1 (Oct4) stained; 5,10: secondary control antibody staining companions to Pou5f1 staining. Note that in F1, H1p70(CM28) is 70 passages total, the last 28 of which were without feeder in CM, in F6, H1p71(B29) is 71 passages total, the last 29 of which were in butyrate and for F7, H1p48(CM3;B6) is 48 passages total, and within the last 9 passages, the first 3 were without feeder in CM and the final 6 passages were in butyrate. This format is followed for all subsequent figures. The size bar indicates 38 µm in C and F1 & 6 and 15 µm in F2–5 &7–10.
Figure 2
Butyrate supports ES cell self-renewal across species. A. H13 cells (1, in CM, 2 in butyrate), rh366.4 rhesus ES cells (3 in CM, 4 in butyrate), R1 mouse ES cells (5 in LIF, 6 in butyrate). B. Cell cycle profile of R1 cells cultured in LIF (left) or butyrate (right). C. BrdU uptake in R1 cells cultured in LIF versus butyrate. ** denotes P<0.01. D. Alkaline phosphatase staining (1, 4), Pou5f1 (Oct4) staining (2, 5), and phase contrast microscopy (3, 6) of R1 cells cultured in LIF (1 – 3) or 0.2 mM butyrate (4 −6). E. Chimeric mouse generated from R1 ES cells cultured for 3 passages in butyrate plus LIF on feeders (panel 1), and its progeny (panel 2), indicating 100% germ line transmission (note all are brown). Chimeric mouse from a mEpiSC line #5, a gift of Paul Tesar and Ron McKay (Tesar et al., 2007) cultured for 18 passages on feeders with the addition of butyrate for the last 8 passages (panel 3). The size bar indicates 38 µm for panels A and D.
Figure 3
Transcriptional responses to butyrate in human and mouse ES cells. Colored dots depict genes that are significantly upregulated (red) or downregulated (green) in response to butyrate in the hESC lines H1 (panels A–D) and BG02 (panel E) and the mESC line R1 (panel F). Panel A - Scatter plot depicting the transcriptional response of H1 cells cultured for 6 passages in butyrate versus H1 cells maintained in CM (see text for details). Panel B - reversion toward the original pattern of expression after returning butyrate treated H1 cells back to CM for 3 passages (“Reconditioned Medium”). Panel C – Scatter plot depicting the transcriptional response to butyrate in BG02 cells (black dots), with red and green dots identifying those genes that were butyrate-regulated in H1 cells, to highlight genes that were coordinately regulated in both hESC lines. Panels D–F contain scatter plots (in black) depicting average expression levels in mEpiSCs versus mESC (from Tesar et al., 2007 - identical for all three panels). Panels D and E overlay butyrate responsive homologous genes in H1 and BG02 cells, respectively. Colored dots indicate homologous genes in the hESC lines that were significantly upregulated (red) or downregulated (green) in response to butyrate. Panel F overlays butyrate responsive genes in mESCs. Colored dot overlays indicate genes in mESCs (R1 cells) that were significantly upregulated (red) or downregulated (green) in response to butyrate. Note that butyrate pulls the gene expression profile of hESCs toward mESCs (X axis) and away from mEpiSCs (Y axis), while pushing mESCs toward EpiSCs, thus the relative orientation of red and green dots between panels D and E versus F is reversed. T-tests indicate that these changes are highly significant (Panel D: t=-9.921, panel E: t=-4.88, panel F: t=11.139, all p<0.001, see Supplemental Experimental Procedures).
Figure 4
Differences in expression levels of 87 ES cell-related genes between H1 cells cultured in butyrate [H1p48(CM3;B6)] versus CM [H1p48(CM9)] (butyrate/CM – orange bars) and mESC versus EpiSCs (ES/EpiSC – green bars from Tesar et al 2007). Y axis indicates Log 2 fold change. Spearman's rho correlation coefficient for the 2 data sets is 0.42, P< 10−4.
Figure 5
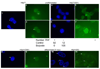
Butyrate cultures are associated with a lack of Xist. Dapi nuclear stain indicating the presumptive presence of the condensed X chromatin (arrows, 1) and accompanying Xist expression (arrows 2 & 3) in later passage H9 cells. The same cells grown for the last 31 passages in butyrate on feeders did not show condensation of the X chromatin (4 and 6) or Xist (5 and 7). Below panels 1–7 are the corresponding counts of Xist positive versus negative cells on the coverslips. Dapi staining of earlier passage H9 cells grown on feeders with one passage in CM shows evidence of X-inactivation upon differentiation for 21 days (8, Dapi and 9, Xist). Cells grown for 3 passages in butyrate followed by differentiation for 21 days in the absence of butyrate showed clear evidence of appropriate Xist body formation induced by differentiation (10, Dapi and 11, Xist). Arrows depict location of condensed chromatin (Dapi) and Xist bodies (green Xist). The size bar indicates 5 µm.
Figure 6
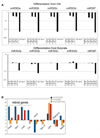
hESCs cultured in butyrate differentiate more slowly than hESCs cultured in CM. A. Time course of 302 family member miRNA expression in H1 cells during an 8-day course of differentiation. Top panels: H1 cells cultured in CM prior to differentiation [H1p69(CM14)]. Bottom panels: H1 cells cultured in butyrate prior to differentiation [H1p67(B15)]. Note that butyrate treated H1 cells exhibit a slower decline in 302 family miRNAs with differentiation compared to H1 cells cultured in CM. B. qRTPCR of differentiation associated transcripts in H1 cells cultured according to a previously published neuroretinal differentiation protocol (Lamba et al., 2006). Blue bars: H1 cells cultured in CM for 8 passages; orange bars: H1 cells cultured in butyrate for 8 passages; red bars: H1 cells cultured in butyrate for 6 passages and reverted to CM for 3 passages. Pax6, Six3, Lhx2, Crx are associated with retinal differentiation.
Figure 7
Epigenetic responses to butyrate. A. H3K9 acetylation in the promoters of butyrate-responsive genes. Light grey bars indicate BG02 cells cultured in CM [BG02p74(CM35)]; dark grey bars indicate BG02 cells cultured in butyrate [BG02p79(CM29;B24)]. An antibody directed against total histone H3 provided a control. B. Bisulfite sequencing of the Dppa5 promoter in H1 cells (left) and BG02 cells (right). Closed circles indicate the presence of methylation and open circles the absence of methylation. Numbers indicate the distance upstream from the transcription start site. Comparisons were made between H1p77(CM43) vs. H1p84(B42) (left panels) and BG02p49(CM10) vs. BG02p76(CM29;B18) (right panels). C. Changes in Dppa5 promoter methylation at various time points following a switch from CM [H1p63(CM8)] to butyrate. Open circles indicate methylation levels in butyrate treated cells whereas the closed circle depicts Dppa5 promoter methylation in the same cells continuously cultured in CM.
Similar articles
- Inhibition of HDAC activity directly reprograms murine embryonic stem cells to trophoblast stem cells.
Huang B, Peng X, Zhai X, Hu J, Chen J, Yang S, Huang Q, Deng E, Li H, Barakat TS, Chen J, Pei D, Fan X, Chambers I, Zhang M. Huang B, et al. Dev Cell. 2024 Aug 19;59(16):2101-2117.e8. doi: 10.1016/j.devcel.2024.05.009. Epub 2024 May 31. Dev Cell. 2024. PMID: 38823394 - Histone deacetylase inhibitor promotes differentiation of embryonic stem cells into neural cells in adherent monoculture.
Yao X, Zhang JR, Huang HR, Dai LC, Liu QJ, Zhang M. Yao X, et al. Chin Med J (Engl). 2010 Mar 20;123(6):734-8. Chin Med J (Engl). 2010. PMID: 20368096 - Inhibition of histone deacetylase activity by butyrate.
Davie JR. Davie JR. J Nutr. 2003 Jul;133(7 Suppl):2485S-2493S. doi: 10.1093/jn/133.7.2485S. J Nutr. 2003. PMID: 12840228 Review. - Small Molecules that Promote Self-Renewal of Stem Cells and Somatic Cell Reprogramming.
Chen G, Guo Y, Li C, Li S, Wan X. Chen G, et al. Stem Cell Rev Rep. 2020 Jun;16(3):511-523. doi: 10.1007/s12015-020-09965-w. Stem Cell Rev Rep. 2020. PMID: 32185667 Review.
Cited by
- The impact of selective HDAC inhibitors on the transcriptome of early mouse embryos.
Shao R, Suzuki T, Suyama M, Tsukada Y. Shao R, et al. BMC Genomics. 2024 Feb 5;25(1):143. doi: 10.1186/s12864-024-10029-3. BMC Genomics. 2024. PMID: 38317092 Free PMC article. - Pluripotency-Associated microRNAs in Early Vertebrate Embryos and Stem Cells.
Maraghechi P, Aponte MTS, Ecker A, Lázár B, Tóth R, Szabadi NT, Gócza E. Maraghechi P, et al. Genes (Basel). 2023 Jul 12;14(7):1434. doi: 10.3390/genes14071434. Genes (Basel). 2023. PMID: 37510338 Free PMC article. Review. - Sinhcaf-dependent histone deacetylation is essential for primordial germ cell specification.
Tao B, Hu H, Chen J, Chen L, Luo D, Sun Y, Ge F, Zhu Z, Trudeau VL, Hu W. Tao B, et al. EMBO Rep. 2022 Jun 7;23(6):e54387. doi: 10.15252/embr.202154387. Epub 2022 May 9. EMBO Rep. 2022. PMID: 35532311 Free PMC article. - Translational and post-translational control of human naïve versus primed pluripotency.
Chen C, Zhang X, Wang Y, Chen X, Chen W, Dan S, She S, Hu W, Dai J, Hu J, Cao Q, Liu Q, Huang Y, Qin B, Kang B, Wang YJ. Chen C, et al. iScience. 2021 Dec 17;25(1):103645. doi: 10.1016/j.isci.2021.103645. eCollection 2022 Jan 21. iScience. 2021. PMID: 35005567 Free PMC article. - Panobinostat Effectively Increases Histone Acetylation and Alters Chromatin Accessibility Landscape in Canine Embryonic Fibroblasts but Does Not Enhance Cellular Reprogramming.
Moshref M, Questa M, Lopez-Cervantes V, Sears TK, Greathouse RL, Crawford CK, Kol A. Moshref M, et al. Front Vet Sci. 2021 Sep 29;8:716570. doi: 10.3389/fvets.2021.716570. eCollection 2021. Front Vet Sci. 2021. PMID: 34660761 Free PMC article.
References
- Abbondanzo SJ, Gadi I, Stewart CL. Derivation of embryonic stem cell lines. Meth Enzymol. 1993;225:803–823. - PubMed
- Andäng M, Hierling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, Pozas E, Brvia V, Halliez S, Nishimaru H, Wilbertz J, Arenas E, Koltzenburg M, Charnay P, El Manira A, Ibañez CF, Ernfors P. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. - PubMed
- Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007;25:207–215. - PubMed
- Becker KA, Stein JL, LIan JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J. Cell Physiol. 2007;210:517–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 GM069983-01/GM/NIGMS NIH HHS/United States
- P20 GM069983-04/GM/NIGMS NIH HHS/United States
- P01 GM081619-020001/GM/NIGMS NIH HHS/United States
- GM076990/GM/NIGMS NIH HHS/United States
- P20 GM069983/GM/NIGMS NIH HHS/United States
- R01 GM076990/GM/NIGMS NIH HHS/United States
- P20 GM069983-010001/GM/NIGMS NIH HHS/United States
- P01 GM081619-010001/GM/NIGMS NIH HHS/United States
- P01 GM081619-02/GM/NIGMS NIH HHS/United States
- P01 GM081619-01/GM/NIGMS NIH HHS/United States
- P01 GM081619/GM/NIGMS NIH HHS/United States
- P20 GM069983-03/GM/NIGMS NIH HHS/United States
- 1P01GM081619/GM/NIGMS NIH HHS/United States
- P20 GM069983-02/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources