Harmonics of circadian gene transcription in mammals - PubMed (original) (raw)
Harmonics of circadian gene transcription in mammals
Michael E Hughes et al. PLoS Genet. 2009 Apr.
Abstract
The circadian clock is a molecular and cellular oscillator found in most mammalian tissues that regulates rhythmic physiology and behavior. Numerous investigations have addressed the contribution of circadian rhythmicity to cellular, organ, and organismal physiology. We recently developed a method to look at transcriptional oscillations with unprecedented precision and accuracy using high-density time sampling. Here, we report a comparison of oscillating transcription from mouse liver, NIH3T3, and U2OS cells. Several surprising observations resulted from this study, including a 100-fold difference in the number of cycling transcripts in autonomous cellular models of the oscillator versus tissues harvested from intact mice. Strikingly, we found two clusters of genes that cycle at the second and third harmonic of circadian rhythmicity in liver, but not cultured cells. Validation experiments show that 12-hour oscillatory transcripts occur in several other peripheral tissues as well including heart, kidney, and lungs. These harmonics are lost ex vivo, as well as under restricted feeding conditions. Taken in sum, these studies illustrate the importance of time sampling with respect to multiple testing, suggest caution in use of autonomous cellular models to study clock output, and demonstrate the existence of harmonics of circadian gene expression in the mouse.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
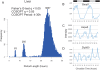
Figure 1. High resolution profiling in the liver identified circadian and sub-ciradian rhythms.
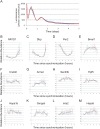
Liver samples were collected every hour for 48 h and analyzed with Affymetrix expression arrays. Rhythmic genes were identified using both COSOPT and Fisher's G-test at a false-discovery rate of <0.05. The period length of every rhythmic transcript was plotted as a histogram; clusters of rhythmic genes with period lengths of approximately 24 (>20 and <30 hours), 12 (>10 and <14 hours) and 8-hours (>7 and <9) were observed (A). In panels B–D, the microarray intensity from three examples was plotted against CT time. Bmal1 (B), Hspa5 (C), and Zfp560 (D) expression profiles demonstrate 24, 12, and 8 h period lengths, respectively.
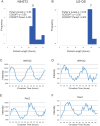
Figure 2. High temporal resolution profiling of NIH3T3 and U2OS cells reveals severely dampened circadian output.
NIH3T3 and U2OS cells were grown to confluence and shocked with either forskolin (NIH3T3) or dexamethasone (U2OS) to synchronize their circadian clocks. mRNA samples were collected every h for 48 h and profiled on Affymetrix expression arrays. Rhythmic genes were identified using both COSOPT and Fisher's G-test at a false-discovery rate of <0.05. The period length of every rhythmic transcript was plotted as a histogram (A–B). To demonstrate that core clock genes cycle well in these data sets, panels C–F show the microarray intensity from two representative genes was plotted against CT time for both NIH3T3 and U2OS cells. NR1D2 (C–D) and Per3 (E–F) expression profiles show examples of cycling 24 h genes.
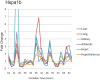
Figure 3. qPCR profiling of Hspa1b reveals 12 h rhythms in multiple tissues.
RNA samples from six different tissues were collected at a two-hour resolution between CT18 and CT64. These samples were analyzed using qPCR probes, median normalized, and plotted against CT time. Notably, in every tissue tested, Hspa1b shows four peaks of expression during the 48 h time course; in every case, the phase of these rhythms is invariant between tissues.
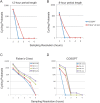
Figure 4. High temporal resolution is required to detect 12 and 8 h rhythms.
COSOPT and Fisher's G-test were performed on subsets of the microarray data set to simulate the statistical power of sampling at a 1, 2, 3, 4 or 6 h resolution. The number of rhythmic genes detected at a FDR of <0.05 by either algorithm is plotted against the sampling resolution for 12 h (period >10 and <14) (A) or 8 h genes (period >7 and <9) (B). In each case, one hour sampling resolution is required to optimally detect transcriptional rhythms. Additional simulations revealed that both Fisher's G test (C) and COSOPT (D) detected considerably more rhythmic transcripts of all period lengths when samples were taken at a two-hour resolution or better. At very high FDRs (e.g. <0.4), using single algorithms, a sizable proportion of the genome is found to cycle. This observation underscores the importance of using appropriately low FDRs as well as multiple algorithms to cross-validate cycling genes.
Figure 5. The peaks of 12 h cycling genes correlate with subjective dusk and dawn.
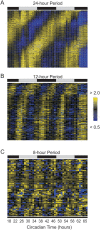
Rhythmic transcripts detected by COSOPT and Fisher's G-test at a false-discovery rate of <0.05 were median-normalized and plotted as a heatmap for 24, 12 and 8 h cycling genes (A–C). Bright yellow represents expression 2-fold greater than median levels while bright blue represents expression less than 50 percent of median levels. The time of peak expression of 24 h cycling genes show a roughly equal distribution over the course of a day; in contrast, the peak expression of both 12 h rhythms are biased to specific times each day.
Figure 6. 12 h rhythmic transcription is dampened in ex vivo hepatocytes.
Primary hepatocytes were prepared from Per2-luciferase mice and shocked with dexamethasone to synchronize their circadian clocks. Real-time luciferase measurements revealed a circadian oscillation which dampens over the course of three days in vitro for two replicates shown in red and blue (A). Starting four h after dexamethasone shock, mRNA samples from these cells were collected every two h for an entire day and quantitative PCR was used to assess the levels of endogenous mRNAs. Core clock genes, including NR1D1, Dbp, Per2 and Bmal1, were rhythmic over the analyzed time points (B–E); however, 12 h genes were either severely dampened (F) or were completely arrhythmic (G–M). Error bars are +/−S.E.M.; thick purple traces represent the average of three replicates, thin traces show the result of each individual replicate.
Figure 7. Restricted feeding changes the periodicity of 12 h rhythms.
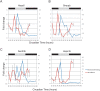
Mice were held in a restricted feeding paradigm (see Methods) and liver samples were collected every 2 h. Quantitative PCR was used to assess the transcriptional profile of eight genes: Hspa5 (A), Gmppb (B), Sec23b (C), Hspa1b (D), as well as Gramd3, Creld2, Gosr2 and Ints2 (data not shown). When compared to samples from ad libitum fed mice (red traces, right axis), restricted feeding samples (blue traces, left axis) showed only a single peak of expression over the course of a complete day (Error bars are +/−S.E.M.).
Similar articles
- Coordinated transcription of key pathways in the mouse by the circadian clock.
Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Panda S, et al. Cell. 2002 May 3;109(3):307-20. doi: 10.1016/s0092-8674(02)00722-5. Cell. 2002. PMID: 12015981 - Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression.
Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Vollmers C, et al. Proc Natl Acad Sci U S A. 2009 Dec 15;106(50):21453-8. doi: 10.1073/pnas.0909591106. Epub 2009 Nov 25. Proc Natl Acad Sci U S A. 2009. PMID: 19940241 Free PMC article. - Transcriptional oscillation of canonical clock genes in mouse peripheral tissues.
Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, Takumi T. Yamamoto T, et al. BMC Mol Biol. 2004 Oct 9;5:18. doi: 10.1186/1471-2199-5-18. BMC Mol Biol. 2004. PMID: 15473909 Free PMC article. - Daily oscillations in liver function: diurnal vs circadian rhythmicity.
Davidson AJ, Castañón-Cervantes O, Stephan FK. Davidson AJ, et al. Liver Int. 2004 Jun;24(3):179-86. doi: 10.1111/j.1478-3231.2004.00917.x. Liver Int. 2004. PMID: 15189266 Review. - Circadian transcriptional output in the SCN and liver of the mouse.
Hogenesch JB, Panda S, Kay S, Takahashi JS. Hogenesch JB, et al. Novartis Found Symp. 2003;253:171-80; discussion 52-5, 102-9, 180-3 passim. Novartis Found Symp. 2003. PMID: 14712921 Review.
Cited by
- Daily rhythms of glycerophospholipid synthesis in fibroblast cultures involve differential enzyme contributions.
Acosta-Rodríguez VA, Márquez S, Salvador GA, Pasquaré SJ, Gorné LD, Garbarino-Pico E, Giusto NM, Guido ME. Acosta-Rodríguez VA, et al. J Lipid Res. 2013 Jul;54(7):1798-811. doi: 10.1194/jlr.M034264. Epub 2013 May 2. J Lipid Res. 2013. PMID: 23641021 Free PMC article. - Assembly of a comprehensive regulatory network for the mammalian circadian clock: a bioinformatics approach.
Lehmann R, Childs L, Thomas P, Abreu M, Fuhr L, Herzel H, Leser U, Relógio A. Lehmann R, et al. PLoS One. 2015 May 6;10(5):e0126283. doi: 10.1371/journal.pone.0126283. eCollection 2015. PLoS One. 2015. PMID: 25945798 Free PMC article. - MetaCycle: an integrated R package to evaluate periodicity in large scale data.
Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. Wu G, et al. Bioinformatics. 2016 Nov 1;32(21):3351-3353. doi: 10.1093/bioinformatics/btw405. Epub 2016 Jul 4. Bioinformatics. 2016. PMID: 27378304 Free PMC article. - Temporal dynamics and composition of ocular surface microbiota in C57BL/6J mice: uncovering a 12h ultradian rhythm.
Jiao X, Li Z. Jiao X, et al. Front Cell Infect Microbiol. 2023 Nov 2;13:1244454. doi: 10.3389/fcimb.2023.1244454. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38029247 Free PMC article. - Estimating confidence intervals in predicted responses for oscillatory biological models.
St John PC, Doyle FJ 3rd. St John PC, et al. BMC Syst Biol. 2013 Jul 29;7:71. doi: 10.1186/1752-0509-7-71. BMC Syst Biol. 2013. PMID: 23895261 Free PMC article.
References
- Curtis AM, Fitzgerald GA. Central and peripheral clocks in cardiovascular and metabolic function. Ann Med. 2006;38:552–559. - PubMed
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. - PubMed
- Klerman EB. Clinical aspects of human circadian rhythms. J Biol Rhythms. 2005;20:375–386. - PubMed
- Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. - PubMed
Publication types
MeSH terms
Grants and funding
- P50 MH074924/MH/NIMH NIH HHS/United States
- R01 NS054794/NS/NINDS NIH HHS/United States
- 1R01NS054794/NS/NINDS NIH HHS/United States
- P50 MH074924-01/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases