Diversity and morphology of members of the phylum "synergistetes" in periodontal health and disease - PubMed (original) (raw)
Diversity and morphology of members of the phylum "synergistetes" in periodontal health and disease
S R Vartoukian et al. Appl Environ Microbiol. 2009 Jun.
Abstract
Members of the phylum "Synergistetes" have frequently been detected in the human oral cavity at sites of dental disease, but they have rarely been detected in studies of oral health. Only two oral "Synergistetes" taxa are cultivable. The aims of this study were to investigate the diversity of "Synergistetes" in the oral cavity, to establish whether "Synergistetes" taxa are more strongly associated with periodontitis than with oral health, and to visualize unculturable "Synergistetes" in situ. Sixty samples (saliva, dental plaque, and mucosal swabs) were collected from five subjects with periodontitis and five periodontally healthy controls. Using phylum-specific 16S rRNA gene primers, "Synergistetes" were identified by PCR, cloning, and sequencing of 48 clones per PCR-positive sample. Subgingival plaque samples were labeled with probes targeting rRNA of unculturable oral "Synergistetes" using fluorescent in situ hybridization (FISH). Analysis of 1,664 clones revealed 12 "Synergistetes" operational taxonomic units (OTUs) at the 99% sequence identity level, 5 of which were novel. "Synergistetes" OTU 4.2 was found in significantly more subjects with periodontitis than controls (P = 0.048) and was more abundant in subgingival plaque at diseased sites than at healthy sites in subjects with periodontitis (P = 0.019) or controls (P = 0.019). FISH analysis revealed that unculturable oral "Synergistetes" cells were large curved bacilli. The human oral cavity harbors a diverse population of "Synergistetes." "Synergistetes" OTU 4.2 is associated with periodontitis and may have a pathogenic role.
Figures
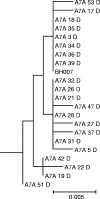
FIG. 1.
Phylogenetic tree based on 16S rRNA gene sequence comparisons, showing the relationships between novel “_Synergistetes_” taxa identified in this study and related species. The tree was constructed using the neighbor-joining method following distance analysis of aligned sequences using the Jukes-Cantor correction and was rooted with Treponema socranskii (accession no. AF033307). The novel sequences were around 750 bp long and were compared in a pairwise manner with virtually full-length sequences of reference strains with unaligned bases for each pair deleted. The numbers at the nodes are bootstrap values for the branches based on data for 500 trees. The accession number for the 16S rRNA sequence is given for each strain. The scale bar indicates 0.05 nucleotide substitutions per site.
FIG. 2.
Phylogenetic tree based on 16S rRNA gene sequence comparisons, showing the variation among representatives of “_Synergistetes_” OTU 3.3 in a single sample (saliva from subject A7A). The tree was constructed using the neighbor-joining method following distance analysis of aligned sequences using the Jukes-Cantor correction, was rooted with “_Synergistetes_” phylotype D084, and was based on 435 unambiguously aligned bases.
FIG. 3.
Detection of 12 “_Synergistetes_” OTUs in cases and controls.
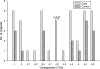
FIG. 4.
Percentage of clones representing each “_Synergistetes_” OTU as a proportion of the total “_Synergistetes_” clones from all sites for cases and controls.
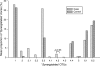
FIG. 5.
Percentage of clones representing each “_Synergistetes_” OTU in subgingival plaque samples from cases and controls.
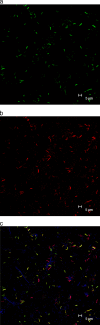
FIG. 6.
Confocal FISH micrographs of subgingival plaque showing (a) “_Synergistetes_” OTU 3.3 (green) (probe 3.3_65 and Alexa Fluor 488), (b) cluster A “_Synergistetes_” (red) (probe A_487 and Cy3), and (c) an overlay for “_Synergistetes_” OTU 3.3 (yellow), cluster A “_Synergistetes_” (yellow and pink), and total bacteria (blue) (probe EUB338 and Cy5).
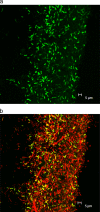
FIG. 7.
FISH images of subgingival plaque showing (a) “_Synergistetes_” OTU 3.3 (green) (probe 3.3_65 and Cy3) and (b) an overlay of “_Synergistetes_” OTU 3.3 (yellow or green) with total bacteria (red) (probe EUB338 and Cy5).
Similar articles
- Distributions of Synergistetes in clinically-healthy and diseased periodontal and peri-implant niches.
Yu XL, Chan Y, Zhuang LF, Lai HC, Lang NP, Lacap-Bugler DC, Leung WK, Watt RM. Yu XL, et al. Microb Pathog. 2016 May;94:90-103. doi: 10.1016/j.micpath.2015.11.029. Epub 2015 Dec 11. Microb Pathog. 2016. PMID: 26686411 - Prevalence and diversity of Synergistetes taxa in periodontal health and disease.
You M, Mo S, Watt RM, Leung WK. You M, et al. J Periodontal Res. 2013 Apr;48(2):159-68. doi: 10.1111/j.1600-0765.2012.01516.x. Epub 2012 Aug 6. J Periodontal Res. 2013. PMID: 22881378 - In-depth snapshot of the equine subgingival microbiome.
Gao W, Chan Y, You M, Lacap-Bugler DC, Leung WK, Watt RM. Gao W, et al. Microb Pathog. 2016 May;94:76-89. doi: 10.1016/j.micpath.2015.11.002. Epub 2015 Nov 10. Microb Pathog. 2016. PMID: 26550763 - Phylum Synergistetes in the oral cavity: A possible contributor to periodontal disease.
McCracken BA, Nathalia Garcia M. McCracken BA, et al. Anaerobe. 2021 Apr;68:102250. doi: 10.1016/j.anaerobe.2020.102250. Epub 2020 Aug 11. Anaerobe. 2021. PMID: 32791127 Review. - Subgingival biofilm structure.
Zijnge V, Ammann T, Thurnheer T, Gmür R. Zijnge V, et al. Front Oral Biol. 2012;15:1-16. doi: 10.1159/000329667. Epub 2011 Nov 11. Front Oral Biol. 2012. PMID: 22142954 Review.
Cited by
- Oral biofilms: molecular analysis, challenges, and future prospects in dental diagnostics.
Do T, Devine D, Marsh PD. Do T, et al. Clin Cosmet Investig Dent. 2013 Feb 28;5:11-9. doi: 10.2147/CCIDE.S31005. Print 2013. Clin Cosmet Investig Dent. 2013. PMID: 23674928 Free PMC article. - In vitro culture of previously uncultured oral bacterial phylotypes.
Thompson H, Rybalka A, Moazzez R, Dewhirst FE, Wade WG. Thompson H, et al. Appl Environ Microbiol. 2015 Dec;81(24):8307-14. doi: 10.1128/AEM.02156-15. Epub 2015 Sep 25. Appl Environ Microbiol. 2015. PMID: 26407883 Free PMC article. - Biogeography of a human oral microbiome at the micron scale.
Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Mark Welch JL, et al. Proc Natl Acad Sci U S A. 2016 Feb 9;113(6):E791-800. doi: 10.1073/pnas.1522149113. Epub 2016 Jan 25. Proc Natl Acad Sci U S A. 2016. PMID: 26811460 Free PMC article. - Armed to the Teeth-The Oral Mucosa Immunity System and Microbiota.
Ptasiewicz M, Grywalska E, Mertowska P, Korona-Głowniak I, Poniewierska-Baran A, Niedźwiedzka-Rystwej P, Chałas R. Ptasiewicz M, et al. Int J Mol Sci. 2022 Jan 14;23(2):882. doi: 10.3390/ijms23020882. Int J Mol Sci. 2022. PMID: 35055069 Free PMC article. Review. - The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems.
Willis JR, Gabaldón T. Willis JR, et al. Microorganisms. 2020 Feb 23;8(2):308. doi: 10.3390/microorganisms8020308. Microorganisms. 2020. PMID: 32102216 Free PMC article. Review.
References
- Beikler, T., G. Abdeen, S. Schnitzer, S. Salzer, B. Ehmke, A. Heinecke, and T. F. Flemmig. 2004. Microbiological shifts in intra- and extraoral habitats following mechanical periodontal therapy. J. Clin. Periodontol. 31:777-783. - PubMed
- Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases