Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations - PubMed (original) (raw)
Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations
Carles Ubeda et al. Mol Microbiol. 2009 Apr.
Abstract
SaPI1 and SaPIbov1 are chromosomal pathogenicity islands in Staphylococcus aureus that carry tst and other superantigen genes. They are induced to excise and replicate by certain phages, are efficiently encapsidated in SaPI-specific small particles composed of phage virion proteins and are transferred at very high frequencies. In this study, we have analysed three SaPI genes that are important for the phage-SaPI interaction, int (integrase) terS (phage terminase small subunit homologue) and pif (phage interference function). SaPI1 int is required for SaPI excision, replication and packaging in a donor strain, and is required for integration in a recipient. A SaPI1 int mutant, following phage induction, produces small SaPI-specific capsids which are filled with partial phage genomes. SaPIbov1 DNA is efficiently packaged into full-sized phage heads as well as into SaPI-specific small ones, whereas SaPI1 DNA is found almost exclusively in the small capsids. TerS, however, determines DNA packaging specificity but not the choice of large versus small capsids. This choice is influenced by SaPIbov1 gene 12, which prevents phage DNA packaging into small capsids, and which is also primarily responsible for interference by SaPIbov1 with phage reproduction.
Figures
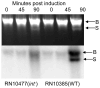
Fig. 1. Effect of int mutation on the SaPI1 ERP cycle
80α lysogens containing either SaPI1 int::tetM (at left) or SaPI1 WT (at right) were mitomycin induced and 5 ml samples removed for DNA analysis at 0, 45, and 90 min. These were used to prepare screening lysates that were analyzed by agarose gel electrophoresis and stained with ethidium bromide or Southern blotted with a SaPI1 probe. B – bulk DNA; S – SaPI1 monomer-sized DNA.
Fig. 2. Electron micrographs of negatively stained phage and SaPI particles
φ80α lysogens were MC induced, then incubated in CY broth until lysis. Lysates were centrifuged to remove debris, then filtered and the filtrates pelleted by centrifugation, allowed to resuspend in phage buffer and analyzed by electron microscopy. A – 80α alone; B – 80α-SaPI1 WT; C – 80α-SaPI1-int::tetM; D – 80α-SaPIbov1 DterSSaPIbov1. L – mature (large) phage particle; S – Mature SaPI (small) particle; LP – phage pro-capsid; SP – SaPI pro-capsid.
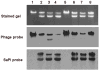
Fig. 3. Packaging specificity
φ80α lysogens were induced, then incubated in CY broth until lysis. DNA extracted from phage particles purified by centrifugation, was analyzed by gel electrophoresis and gel was blotted with a SaPI-specific probe or phage-specific probes downstream or upstream of the 80α pac site. Composition of the strains used for the preparation of lysates is indicated above.
Fig. 4. Effects of SaPIbov1 mutations on SaPIbov1 and phage DNA packaging
Samples were prepared and analyzed as in Fig. 3. 1- φ80α; 2- φ80α SaPIbov1 wt; 3- φ80α SaPIbov1 Δ gene 12; 4- φ80α SaPIbov1 Δ gene 12 + pCN51:gene12; 5- φ80α SaPIbov1 Δ ter; 6- φ80α SaPIbov1 Δ gene 6; 7- φ80α SaPIbov1 Δ gene 10; 8- φ80α SaPIbov1 Δ gene 11;
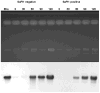
Fig. 5. Effect of SaPI1 on 80α replication
Cultures of RN10616 (φ80α, SaPI1-) or RN10628 (φ80α, SaPI1+) were grown to a Klett=30, UV-induced, and aliquots were removed at the indicated times and used to prepare minilysates. Equivalent amounts of total DNA were loaded in each well of an agarose gel and separated by electrophoresis. Gels were stained and photographed, and then transferred and probed with an 80α probe from the replication region (generated with primers SMT98 and SMT99). The upper panel shows the stained gel; the lower panel is an autoradiogram of the Southern blot. The first lane contains purified 80α genomic DNA. Similar results were obtained following 80α infection of a SaPI1-negative and SaPI1-positive strain (data not shown). P- phage monomer-sized band; S - SaPI1 monomer-sized band.
Fig. 6. Effect of cloned SaPIbov1 GP12 on 80α DNA replication and packaging
Cultures of RN4220 derivatives were infected with 80α at a multiplicity of 3 and incubated in CY broth for 1 h, at which time, mini-lysates were prepared and separated on agarose. The gels were stained and photographed (left panel), then Southern blotted (right panel) with an 80α c1 gene probe. Bulk – phage and chromosomal DNAs; SaPI – SaPI-sized monomers released from intracellular phage capsids; Lane 1, pCN51 (vector); 2, pCN51::gene12; 3, SaPIbov1Δ12 + pCN51; 4, SaPIbov1Δ12 + pCN51::gene12; 5, SaPIbov1; 6, SaPI1 + pCN51; 7, SaPI1 + pCN51::gene12. Cultures used for lower gel were grown with 0.1 mM CdCl2.
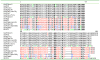
Fig. 7. Homologs of SaPIbov1 GP12 (phage interference function)
Residues defining the SaPI1 group of Pifs are mostly in red. In blue are residues in SaPI5 and SaPImw2 that are specifically different from the other SaPIs. Residues shown in green differ from consensus sequences in undefined ways.
Similar articles
- Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer.
Maiques E, Ubeda C, Tormo MA, Ferrer MD, Lasa I, Novick RP, Penadés JR. Maiques E, et al. J Bacteriol. 2007 Aug;189(15):5608-16. doi: 10.1128/JB.00619-07. Epub 2007 Jun 1. J Bacteriol. 2007. PMID: 17545290 Free PMC article. - Sequence determinants for DNA packaging specificity in the S. aureus pathogenicity island SaPI1.
Bento JC, Lane KD, Read EK, Cerca N, Christie GE. Bento JC, et al. Plasmid. 2014 Jan;71:8-15. doi: 10.1016/j.plasmid.2013.12.001. Epub 2013 Dec 21. Plasmid. 2014. PMID: 24365721 Free PMC article. - The complete genomes of Staphylococcus aureus bacteriophages 80 and 80α--implications for the specificity of SaPI mobilization.
Christie GE, Matthews AM, King DG, Lane KD, Olivarez NP, Tallent SM, Gill SR, Novick RP. Christie GE, et al. Virology. 2010 Nov 25;407(2):381-90. doi: 10.1016/j.virol.2010.08.036. Epub 2010 Sep 25. Virology. 2010. PMID: 20869739 Free PMC article. - The SaPIs: mobile pathogenicity islands of Staphylococcus.
Novick RP, Subedi A. Novick RP, et al. Chem Immunol Allergy. 2007;93:42-57. doi: 10.1159/000100857. Chem Immunol Allergy. 2007. PMID: 17369699 Review. - Virus DNA packaging: the strategy used by phage lambda.
Catalano CE, Cue D, Feiss M. Catalano CE, et al. Mol Microbiol. 1995 Jun;16(6):1075-86. doi: 10.1111/j.1365-2958.1995.tb02333.x. Mol Microbiol. 1995. PMID: 8577244 Review.
Cited by
- The phage-related chromosomal islands of Gram-positive bacteria.
Novick RP, Christie GE, Penadés JR. Novick RP, et al. Nat Rev Microbiol. 2010 Aug;8(8):541-51. doi: 10.1038/nrmicro2393. Nat Rev Microbiol. 2010. PMID: 20634809 Free PMC article. Review. - A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80α capsids.
Spilman MS, Dearborn AD, Chang JR, Damle PK, Christie GE, Dokland T. Spilman MS, et al. J Mol Biol. 2011 Jan 21;405(3):863-76. doi: 10.1016/j.jmb.2010.11.047. Epub 2010 Dec 1. J Mol Biol. 2011. PMID: 21129380 Free PMC article. - Generalized bacterial genome editing using mobile group II introns and Cre-lox.
Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, Whitt JT, Keatinge-Clay AT, Lambowitz AM, Ellington AD. Enyeart PJ, et al. Mol Syst Biol. 2013;9:685. doi: 10.1038/msb.2013.41. Mol Syst Biol. 2013. PMID: 24002656 Free PMC article. - Noncontiguous operon atlas for the Staphylococcus aureus genome.
Iturbe P, Martín AS, Hamamoto H, Marcet-Houben M, Galbaldón T, Solano C, Lasa I. Iturbe P, et al. Microlife. 2024 Mar 30;5:uqae007. doi: 10.1093/femsml/uqae007. eCollection 2024. Microlife. 2024. PMID: 38651166 Free PMC article. - Structure of the Portal Complex from Staphylococcus aureus Pathogenicity Island 1 Transducing Particles In Situ and In Isolation.
Mukherjee A, Kizziah JL, Hawkins NC, Nasef MO, Parker LK, Dokland T. Mukherjee A, et al. J Mol Biol. 2024 Feb 15;436(4):168415. doi: 10.1016/j.jmb.2023.168415. Epub 2023 Dec 21. J Mol Biol. 2024. PMID: 38135177
References
- Barry P. Microbiology. New York University; 2006. Molecular genetics of the SaPI1 life cycle in Staphylococcus aureus.
- Bowden DW, Modrich P. In vitro maturation of circular bacteriophage P2 DNA. Purification of ter components and characterization of the reaction. J Biol Chem. 1985;260:6999–7007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI022159/AI/NIAID NIH HHS/United States
- R21 AI067654/AI/NIAID NIH HHS/United States
- R21 AI067654-02/AI/NIAID NIH HHS/United States
- R01 AI22159/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources