Sorting of lipids and proteins in membrane curvature gradients - PubMed (original) (raw)
Sorting of lipids and proteins in membrane curvature gradients
A Tian et al. Biophys J. 2009.
Abstract
The sorting of lipids and proteins in cellular trafficking pathways is a process of central importance in maintaining compartmentalization in eukaryotic cells. However, the mechanisms behind these sorting phenomena are currently far from being understood. Among several mechanistic suggestions, membrane curvature has been invoked as a means to segregate lipids and proteins in cellular sorting centers. To assess this hypothesis, we investigate the sorting of lipid analog dye trace components between highly curved tubular membranes and essentially flat membranes of giant unilamellar vesicles. Our experimental findings indicate that intracellular lipid sorting, contrary to frequent assumptions, is unlikely to occur by lipids fitting into membrane regions of appropriate curvature. This observation is explained in the framework of statistical mechanical lattice models that show that entropy, rather than curvature energy, dominates lipid distribution in the absence of strongly preferential lateral intermolecular interactions. Combined with previous findings of curvature induced phase segregation, we conclude that lipid cooperativity is required to enable efficient sorting. In contrast to lipid analog dyes, the peripheral membrane binding protein Cholera toxin subunit B is effectively curvature-sorted. The sorting of Cholera toxin subunit B is rationalized by statistical models. We discuss the implications of our findings for intracellular sorting mechanisms.
Figures
Figure 1
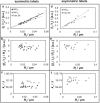
(a) Schematic configuration of a vesicle aspiration and tube pulling experiment, where _P_i, _P_o, and _P_p are the pressures inside vesicle, outside vesicle and in the micropipette, respectively; _R_t, _R_v, and _R_p are the radius of tube, vesicle, and pipette, respectively; _L_t and _L_p are the lengths of tube and vesicle projection in pipette, respectively; f indicates pulling force. (b) Combined transmitted light/confocal fluorescence microscopy image of aspirated vesicle and membrane tether that is pulled by means of a bead. Scale bar = 5 _μ_m. (c) Cross section image of a membrane tether obtained by confocal line scan as indicated by the gray area in panel b. Scale bar = 1 _μ_m. (d) Stepwise pulling and releasing demonstration of tether radius measurements. Tether radius is 44 nm. (e) Tether fluorescence intensity recorded under varying laser power. Tether radius = 22 nm. (f) Molecular structures of DiI derivatives. (g) Molecular structure of CTB (obtained from PDB: 1FGB), lipid-binding sites are at the bottom. A negative spontaneous curvature is suggested.
Figure 2
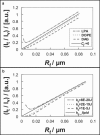
Partitioning of DiI derivatives in POPC membrane tethers. (a_–_c) Plots of double leaflet labeled DiI dyes from the same experiments with shared figure legend shown in a. (d_–_f) Plots of outer leaflet labeled DiI dyes from the same experiments with shared legend shown in d. (a, b, d, and e) Relationship of DiI dye intensities in tethers and tether radius. (a and d) Tether intensities normalized with respect to the intensities of vesicles from which tethers were pulled. Black solid lines, gray solid lines, and black dashed lines are the trend lines of DiIC16, FAST DiI, and DiIC12 data points, respectively. (b and e) Tether intensity to vesicle intensity ratios of panels a and b respectively scaled with respect to corresponding membrane curvature plotted versus tether radius. (c and f) Bending stiffness at varying tether radius.
Figure 3
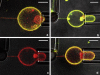
Combined images of red, green and transmitted light channels of POPC (1% GM1) tethers and vesicles coated with CTB. (a) Lipid membrane labeled symmetrically (i.e., in both leaflets) with TR-DHPE, whereas CTB-A488 was added before tether formed; _R_t = 63 nm. (b) Membrane symmetrically labeled with BODIPY-DHPE. CTB-A555 was added before tether formed; _R_t = 66 nm. (c and d) Membrane symmetrically labeled with TR-DHPE; _R_t = 50 nm, images taken before (c) and 18 min after (d) CTB-A488 was added. Scale bar = 5 _μ_m.
Figure 4
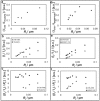
(a and d) POPC tether membrane fluorescence intensity ratios of (a) CTB-A488 to TR-DHPE and (d) CTB-A555 to BODIPY-DHPE (BODIPY-PE) versus tether radius plots of tethers containing (a) 1% GM1 and (d) 3% GM1. (b and e) Normalized tether intensity ratios with respect to vesicle intensities of (b) CTB-A488 and TR-DHPE and (e) CTB-A555 and BODIPY-DHPE at varying tether radius of POPC membranes with 1% GM1 (b) and 3% GM1 (e). (c and f) Same data sets as panels b and e, respectively; curvature-scaled tether intensities plotted with respect to tether radius. (a_–_c) Tether radius was measured from stepwise elongation and releasing as described in Materials and Methods. (d_–_f) Approximate tether radius calculated from membrane tension by assuming _k_c = 8 × 10−20 J.
Figure 5
(a and b) Reversibility test of tether intensity at varying tether radius of POPC (2% GM1) membrane labeled only with CTB-A488. Tether intensity is normalized with respect to vesicle intensity. The dashed line refers to the fitting of data points using the spontaneous curvature sorting model, and the gray solid line to fitting with the bending stiffness sorting model. (b) Same data as in panel a additionally scaled with membrane curvature. (c) Bending stiffness versus tether radius of POPC (1% GM1) membrane labeled with both CTB-A488 and TR-DHPE. Note that these bending stiffness values refer to the data set of the first column in Fig. 4.
Figure 6
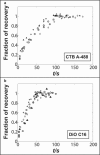
Fluorescence recovery after photobleaching data comparing (a) CTB-A488 and (b) DiOC16(3) on membrane tethers with similar radius ∼65 nm and tether length 19 _μ_m. Whole tethers are bleached in this experiment. Each panel contains five data sets shown with different symbols. CTB is observed to diffuse more slowly compared to DiO on membrane tubes. See the main text for a comparison to diffusion measurements in planar membranes.
Figure 7
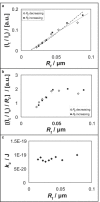
(a) Relative theoretical outer leaflet tether fluorescence intensity as a function of tether radius calculated from spontaneous curvature sorting model, for two extreme spontaneous lipid curvatures (DAG and LPA), or moderate spontaneous curvature (DOPE), as well as a cylindrical (i.e., zero spontaneous curvature) lipid (solid line). Tether intensity is calculated from Eq. 6 (a = 0.5 nm2, T = 295 K). Note that the presence of spontaneous curvature primarily shifts the curves along the ordinate (see Eq. 7). (b) Theoretical tether fluorescence intensity plots calculated from bending stiffness model. Different bending stiffness is applied as legend shows. Tether intensity is calculated based on I∝Rtϕαotϕαov, in which the molar ratios were obtained from Eq. 8 (a = 25 nm2, T = 295 K).
Similar articles
- Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins.
Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, Goud B, Bassereau P. Sorre B, et al. Proc Natl Acad Sci U S A. 2009 Apr 7;106(14):5622-6. doi: 10.1073/pnas.0811243106. Epub 2009 Mar 20. Proc Natl Acad Sci U S A. 2009. PMID: 19304798 Free PMC article. - Lipid cosorting mediated by shiga toxin induced tubulation.
Safouane M, Berland L, Callan-Jones A, Sorre B, Römer W, Johannes L, Toombes GE, Bassereau P. Safouane M, et al. Traffic. 2010 Dec;11(12):1519-29. doi: 10.1111/j.1600-0854.2010.01116.x. Epub 2010 Sep 30. Traffic. 2010. PMID: 20887377 - Curvature sorting of proteins on a cylindrical lipid membrane tether connected to a reservoir.
Singh P, Mahata P, Baumgart T, Das SL. Singh P, et al. Phys Rev E Stat Nonlin Soft Matter Phys. 2012 May;85(5 Pt 1):051906. doi: 10.1103/PhysRevE.85.051906. Epub 2012 May 14. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 23004787 Free PMC article. - Curvature-driven lipid sorting in biomembranes.
Callan-Jones A, Sorre B, Bassereau P. Callan-Jones A, et al. Cold Spring Harb Perspect Biol. 2011 Feb 1;3(2):a004648. doi: 10.1101/cshperspect.a004648. Cold Spring Harb Perspect Biol. 2011. PMID: 21421916 Free PMC article. Review. - Comparing physical mechanisms for membrane curvature-driven sorting of BAR-domain proteins.
Tsai FC, Simunovic M, Sorre B, Bertin A, Manzi J, Callan-Jones A, Bassereau P. Tsai FC, et al. Soft Matter. 2021 Apr 28;17(16):4254-4265. doi: 10.1039/d0sm01573c. Soft Matter. 2021. PMID: 33870384 Review.
Cited by
- Molecular Sensing and Manipulation of Protein Oligomerization in Membrane Nanotubes with Bolaamphiphilic Foldamers.
Aftahy K, Arrasate P, Bashkirov PV, Kuzmin PI, Maurizot V, Huc I, Frolov VA. Aftahy K, et al. J Am Chem Soc. 2023 Nov 22;145(46):25150-25159. doi: 10.1021/jacs.3c05753. Epub 2023 Nov 10. J Am Chem Soc. 2023. PMID: 37948300 Free PMC article. - Molecular structure of membrane tethers.
Baoukina S, Marrink SJ, Tieleman DP. Baoukina S, et al. Biophys J. 2012 Apr 18;102(8):1866-71. doi: 10.1016/j.bpj.2012.03.048. Biophys J. 2012. PMID: 22768942 Free PMC article. - Revisiting Membrane Microdomains and Phase Separation: A Viral Perspective.
Sengupta P, Lippincott-Schwartz J. Sengupta P, et al. Viruses. 2020 Jul 10;12(7):745. doi: 10.3390/v12070745. Viruses. 2020. PMID: 32664429 Free PMC article. Review. - Mode specific elastic constants for the gel, liquid-ordered, and liquid-disordered phases of DPPC/DOPC/cholesterol model lipid bilayers.
Uline MJ, Szleifer I. Uline MJ, et al. Faraday Discuss. 2013;161:177-91; discussion 273-303. doi: 10.1039/c2fd20091k. Faraday Discuss. 2013. PMID: 23805743 Free PMC article. - Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization.
Wu T, Shi Z, Baumgart T. Wu T, et al. PLoS One. 2014 Apr 22;9(4):e93060. doi: 10.1371/journal.pone.0093060. eCollection 2014. PLoS One. 2014. PMID: 24755653 Free PMC article.
References
- Gennis R.B. Springer-Verlag; New York: 1989. Biomembranes: Molecular Structure and Function.
- Holthuis J.C.M., Levine T.P. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 2005;6:209–220. - PubMed
- De Matteis M.A., Luini A. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 2008;9:273–284. - PubMed
- Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources