A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming - PubMed (original) (raw)
doi: 10.1038/ng.358. Epub 2009 Apr 6.
Takashi Sasaki, Aileen Sandilands, Linda E Campbell, Sean P Saunders, Niamh E Mangan, John J Callanan, Hiroshi Kawasaki, Aiko Shiohama, Akiharu Kubo, John P Sundberg, Richard B Presland, Philip Fleckman, Nobuyoshi Shimizu, Jun Kudoh, Alan D Irvine, Masayuki Amagai, W H Irwin McLean
Affiliations
- PMID: 19349982
- PMCID: PMC2872154
- DOI: 10.1038/ng.358
A homozygous frameshift mutation in the mouse Flg gene facilitates enhanced percutaneous allergen priming
Padraic G Fallon et al. Nat Genet. 2009 May.
Abstract
Loss-of-function mutations in the FLG (filaggrin) gene cause the semidominant keratinizing disorder ichthyosis vulgaris and convey major genetic risk for atopic dermatitis (eczema), eczema-associated asthma and other allergic phenotypes. Several low-frequency FLG null alleles occur in Europeans and Asians, with a cumulative frequency of approximately 9% in Europe. Here we report a 1-bp deletion mutation, 5303delA, analogous to common human FLG mutations, within the murine Flg gene in the spontaneous mouse mutant flaky tail (ft). We demonstrate that topical application of allergen to mice homozygous for this mutation results in cutaneous inflammatory infiltrates and enhanced cutaneous allergen priming with development of allergen-specific antibody responses. These data validate flaky tail as a useful model of filaggrin deficiency and provide experimental evidence for the hypothesis that antigen transfer through a defective epidermal barrier is a key mechanism underlying elevated IgE sensitization and initiation of cutaneous inflammation in humans with filaggrin-related atopic disease.
Figures
Figure 1. Murine filaggrin gene structure and mutation identification
(a) The mouse filaggrin gene (flg) resides within a cluster of 7 genes encoding closely related fused-S100 proteins, spanning ~350 kb on murine chromosomal band 3QF21. Like the human ortholog, flg has a 3 exon gene structure. Exon 1 consists of 5’UTR sequences only; exon 2 contains the ATG and encodes part of the N-terminal S100 calcium binding domain of profilaggrin; and exon 3 encodes the remainder of the S100 domain, the B-domain and the filaggrin polyprotein repeat domain. In the July 2007 mouse genome sequence, annotated here (_Flg_C57), the latter consists of 16 near-perfect repeats encoding a 250 amino acid filaggrin protein (1–16) flanked by two imperfect repeats (0 and 17). Repeat 17 is followed by a short tail domain. The 485 bp 3’UTR contains a consensus polyadenylation signal. In ft mice, the exon 3 sequence determined here (Flgft) lacks one repeat due to a 750-bp in-frame deletion, as well as the frameshift mutation c.5303delA. In addition, there are several additional SNPs and small in-frame insertions/deletions in flg that differ between the two strains, so that their DNA sequences overall are 99.3% identical, excluding the repeat copy number variation. (b) Wild-type flg sequence, corresponding to codons 1765–1771, with amino acid translation. The A dinucleotide involved in the mutation is boxed (compare panel c). (c) Analogous flg sequence as shown in (b), derived from a homozygous ft/ft mouse, showing homozygous frameshift mutation designated c.5303delA, predicting truncation mutation p.Asn1768ThrfsX154. The single A nucleotide remaining at the site of the mutation is boxed (compare panel b). (d) Allele-specific genotyping of the parents and F1 offspring of a cross between an ft/ft homozygote and a wild-type mouse. M, molecular weight markers, upper band = 118 bp; lower band = 72 bp. The allele specific band is only amplified from mice carrying the ft mutation. (e) Genotyping for an Acc I restriction site polymorphism in filaggrin repeat 1, approximately 4 kb upstream of the 5303delA mutation. This assay is more convenient for scoring of heterozygotes. The 678 bp C57BL/6 allele does not digest; the ft allele cuts to yield fragments of 559 bp and 134 bp. M, molecular weight markers.
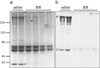
Figure 2. A truncated profilaggrin is expressed in flaky tail mouse skin, which lacks the C-terminus
Urea/Tris protein extracts from control wild type and ft/ft mice were fractionated by SDS/PAGE and immunoblotted with the peptide antibody developed to the C-terminus of mouse profilaggrin. (a) Coomassie brilliant blue-stained protein gel to control for loading, and (b), corresponding immunoblot probed with the C-terminal antibody. The antibody detects both full-length profilaggrin (P) and a putative C-terminal processing product (C) in wild type mouse extracts, however, these immunoreactive products are not detected in ft/ft mice (b). Note the lack of filaggrin (F) in ft/ft mice (a), observed previously. The mutant profilaggrin protein in ft/ft mice is faintly visible in (a) at an apparent molecular weight of ~215 kDa (*). This is readily detectable with a mouse filaggrin antibody as reported previously. Molecular weight marker sizes are shown at left.
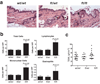
Figure 3. Cutaneous inflammation in untreated ft/ft mice but not wt/wt or wt/ft animals
(a) Representative photomicrographs of skin sections from age- and sex-matched wt/wt, wt/ft and ft/ft mice. Ft/ft mice had orthokeratotic hyperkeratosis (arrow) with occasional foci of acanthosis (bracket) compared to wt/wt and ft/wt mice. (Original magnification x40.) (b). Numbers of skin infiltrating cells, lymphocytes, eosinophils and mononuclear cells were detected per high-power fields (HPF). Cells were counted on 15–20 HPF (x1,000) on hematoxylin and eosin stained sections of 5–6 wt/wt, wt/ft and ft/ft mice. Data represent the mean; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant. (c) TEWL analysis of untreated wt/wt, wt/ft and ft/ft mice. Values are individual mice and Mean bars are shown.
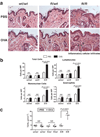
Figure 4. Allergen exposure exacerbates skin inflammation ft/ft mice but not wt/wt or wt/ft animals
(a) Representative photomicrographs of skin sections from age- and sex-matched wt/wt, wt/ft and ft/ft mice exposed to OVA or PBS as a vehicle control. Ft/ft mice had diffuse mild acanthosis and marked increases in dermal cell infiltration when cutaneously challenged with OVA, compared to wt/wt and wt/ft mice. (Original magnification x40.) (b). Quantification of numbers of skin infiltrating cells, lymphocytes, eosinophils and mononuclear cells detected per high-power fields (HPF). Cells were counted at on 15–20 HPF (x1,000) on hematoxylin and eosin stained sections of 11–14 wt/wt, wt/ft and ft/ft mice. Data represent the mean; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant. (c) TEWL analysis of skin of OVA exposed wt/wt, wt/ft and ft/ft mice. Values relate to individual mice and mean bars are shown.
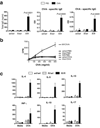
Figure 5. Elevated OVA-specific immune response in allergen exposed ft/ft mice but not wt/wt or wt/ft animals
(a) ELISA detection of levels of total IgE, OVA-specific IgE and IgG in serum from wt/wt, wt/ft and ft/ft mice treated with PBS or OVA on skin. Data are Mean plus Standard Error of the Mean (SEM) from 7–12 mice. (b) Cell proliferation to OVA of spleen cells from wt/wt, wt/ft and ft/ft mice treated with PBS or OVA on skin. Data represent the mean of spleens from 3–4 individual mice per group; error bars represent standard error of the mean. (c) Cytokine production by spleen cells from OVA exposed wt/wt, wt/ft and ft/ft mice treated with PBS or OVA. Cells were cultured in media or OVA (500 µg/ml) and cytokines detected by ELISA. Data represent the mean of spleens from 3–4 individual mice per group; error bars represent standard error of the mean. Student’s t-test, or corrected Welch corrected t-test, was used to determine statistical differences between groups. NS = Non-significant.
Comment in
- Of flaky tails and itchy skin.
Vercelli D. Vercelli D. Nat Genet. 2009 May;41(5):512-3. doi: 10.1038/ng0509-512. Nat Genet. 2009. PMID: 19399034
Similar articles
- Low Threshold for Cutaneous Allergen Sensitization but No Spontaneous Dermatitis or Atopy in FLG-Deficient Mice.
Muhandes L, Chapsa M, Pippel M, Behrendt R, Ge Y, Dahl A, Yi B, Dalpke A, Winkler S, Hiller M, Boutin S, Beissert S, Jessberger R, Fallon PG, Roers A. Muhandes L, et al. J Invest Dermatol. 2021 Nov;141(11):2611-2619.e2. doi: 10.1016/j.jid.2021.02.763. Epub 2021 Apr 22. J Invest Dermatol. 2021. PMID: 33894197 - Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema.
Sandilands A, Terron-Kwiatkowski A, Hull PR, O'Regan GM, Clayton TH, Watson RM, Carrick T, Evans AT, Liao H, Zhao Y, Campbell LE, Schmuth M, Gruber R, Janecke AR, Elias PM, van Steensel MA, Nagtzaam I, van Geel M, Steijlen PM, Munro CS, Bradley DG, Palmer CN, Smith FJ, McLean WH, Irvine AD. Sandilands A, et al. Nat Genet. 2007 May;39(5):650-4. doi: 10.1038/ng2020. Epub 2007 Apr 8. Nat Genet. 2007. PMID: 17417636 - FLG mutations in ichthyosis vulgaris and atopic eczema: spectrum of mutations and population genetics.
Akiyama M. Akiyama M. Br J Dermatol. 2010 Mar;162(3):472-7. doi: 10.1111/j.1365-2133.2009.09582.x. Epub 2009 Dec 2. Br J Dermatol. 2010. PMID: 19958351 Review. - Filaggrin loss-of-function mutations as a predictor for atopic eczema, allergic sensitization and eczema-associated asthma in Polish children population.
Dębińska A, Danielewicz H, Drabik-Chamerska A, Kalita D, Boznański A. Dębińska A, et al. Adv Clin Exp Med. 2017 Sep;26(6):991-998. doi: 10.17219/acem/61430. Adv Clin Exp Med. 2017. PMID: 29068602 - Filaggrin gene defects and the risk of developing allergic disorders.
Osawa R, Akiyama M, Shimizu H. Osawa R, et al. Allergol Int. 2011 Mar;60(1):1-9. doi: 10.2332/allergolint.10-RAI-0270. Epub 2011 Dec 25. Allergol Int. 2011. PMID: 21173567 Review.
Cited by
- When allergies have no name: is idiopathic anaphylaxis driven by co-factors?
Elkhalifa S, Elbashir H, Abuzakouk M. Elkhalifa S, et al. Front Allergy. 2024 Oct 18;5:1468945. doi: 10.3389/falgy.2024.1468945. eCollection 2024. Front Allergy. 2024. PMID: 39493748 Free PMC article. Review. - Preclinical Models of Atopic Dermatitis Suitable for Mechanistic and Therapeutic Investigations.
Maskey AR, Mo X, Li XM. Maskey AR, et al. J Inflamm Res. 2024 Oct 2;17:6955-6970. doi: 10.2147/JIR.S467327. eCollection 2024. J Inflamm Res. 2024. PMID: 39372589 Free PMC article. Review. - Educational and psychological interventions for managing atopic dermatitis (eczema).
Singleton H, Hodder A, Almilaji O, Ersser SJ, Heaslip V, O'Meara S, Boyers D, Roberts A, Scott H, Van Onselen J, Doney L, Boyle RJ, Thompson AR. Singleton H, et al. Cochrane Database Syst Rev. 2024 Aug 12;8(8):CD014932. doi: 10.1002/14651858.CD014932.pub2. Cochrane Database Syst Rev. 2024. PMID: 39132734 Free PMC article. Review. - Early-life risk factors which govern pro-allergic immunity.
Ptaschinski C, Gibbs BF. Ptaschinski C, et al. Semin Immunopathol. 2024 Jul 27;46(3-4):9. doi: 10.1007/s00281-024-01020-x. Semin Immunopathol. 2024. PMID: 39066790 Free PMC article. Review. - Atopic Dermatitis: Disease Background and Risk Factors.
Li B, Fuxench ZC. Li B, et al. Adv Exp Med Biol. 2024;1447:11-19. doi: 10.1007/978-3-031-54513-9_2. Adv Exp Med Biol. 2024. PMID: 38724780
References
- Smith FJD, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. - PubMed
- Palmer CNA, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. - PubMed
- Baurecht H, et al. Toward a major risk factor for atopic eczema: meta-analysis of filaggrin polymorphism data. J Allergy Clin Immunol. 2007;120:1406–1412. - PubMed
- Sandilands A, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007:650–654. - PubMed
- Henderson J, et al. The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J Allergy Clin Immunol. 2008;121:872–877. e9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR049183/AR/NIAMS NIH HHS/United States
- R01 AR49183/AR/NIAMS NIH HHS/United States
- R01 AR049183-01/AR/NIAMS NIH HHS/United States
- G0700314/MRC_/Medical Research Council/United Kingdom
- P01 AM21557/AM/NIADDK NIH HHS/United States
- R01 AR049183-02/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous