Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells - PubMed (original) (raw)
Comparative Study
Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells
Xiangduo Kong et al. Nucleic Acids Res. 2009 May.
Abstract
Proper recognition and repair of DNA damage is critical for the cell to protect its genomic integrity. Laser microirradiation ranging in wavelength from ultraviolet A (UVA) to near-infrared (NIR) can be used to induce damage in a defined region in the cell nucleus, representing an innovative technology to effectively analyze the in vivo DNA double-strand break (DSB) damage recognition process in mammalian cells. However, the damage-inducing characteristics of the different laser systems have not been fully investigated. Here we compare the nanosecond nitrogen 337 nm UVA laser with and without bromodeoxyuridine (BrdU), the nanosecond and picosecond 532 nm green second-harmonic Nd:YAG, and the femtosecond NIR 800 nm Ti:sapphire laser with regard to the type(s) of damage and corresponding cellular responses. Crosslinking damage (without significant nucleotide excision repair factor recruitment) and single-strand breaks (with corresponding repair factor recruitment) were common among all three wavelengths. Interestingly, UVA without BrdU uniquely produced base damage and aberrant DSB responses. Furthermore, the total energy required for the threshold H2AX phosphorylation induction was found to vary between the individual laser systems. The results indicate the involvement of different damage mechanisms dictated by wavelength and pulse duration. The advantages and disadvantages of each system are discussed.
Figures
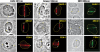
Figure 1.
Induction of different types of DNA damage by UVA, ns and ps green, and NIR lasers. At 3–5 min after damage induction by the different lasers indicated at the top, cells were fixed and stained with antibodies specific for CPD, 6-4PP and 8-oxoG. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm.
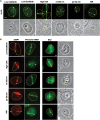
Figure 2.
Recruitment of SSB repair and BER factors to the damage sites induced by UVA, green and NIR lasers. Immediately following the damage induction, cells were fixed and stained with antibodies specific for PARP-1, XRCC1 or FEN-1. The location of the induced lesions are indicated by arrowheads. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm.
Figure 3.
DSB responses induced by different laser systems. (A) Ku recruitment to the laser-induced damage sites. Immunostaining of cells damaged by different lasers as indicated using anti-Ku antibody. Lesions are indicated by arrowheads. Corresponding brightfield phase contrast images are also shown. Scale bar = 5 µm. (B) 53BP1 accumulation, hSMC1 phosphorylation and cohesin accumulation at the damage sites. Cells damaged by different lasers as indicated were fixed and stained with antibodies specific for 53BP1, phosphorylated hSMC1 (S966P), or the non-SMC cohesin subunit SA2. Lesions are indicated by arrowheads. Corresponding brightfield phase contrast images for SA2 staining are also shown. Scale bar = 5 µm.
Figure 4.
Mechanisms of DNA damage by different laser microbeam systems. Three possible damage mechanisms (single-photon absorption, multi-photon absorption, and plasma formation) and their associated thermal, chemical and mechanical effects are listed. Based on the γH2AX-threshold and working laser parameters (e.g. wavelength, pulse duration and frequency, peak irradiance and total input energy), the most likely mechanisms of damage induction by UVA, UVA with BrdU, cw blue, cw blue with BrdU, ns and ps green, and fs NIR are indicated. Although the thermal effect is most likely an important mechanism of damage induction, it is highly restricted temporally and spatially, and is dispersed instantaneously, and therefore any heat-inflicted damage outside of the focused area or any temperature rise in the cell is not expected to occur under the conditions used. Further study is necessary to delineate which mechanisms and/or combination of mechanisms impact each of the laser and biological systems discussed.
Similar articles
- Use of near infrared femtosecond lasers as sub-micron radiation microbeam for cell DNA damage and repair studies.
Botchway SW, Reynolds P, Parker AW, O'Neill P. Botchway SW, et al. Mutat Res. 2010 Apr-Jun;704(1-3):38-44. doi: 10.1016/j.mrrev.2010.01.003. Epub 2010 Jan 14. Mutat Res. 2010. PMID: 20079460 Review. - In situ analysis of DNA damage response and repair using laser microirradiation.
Kim JS, Heale JT, Zeng W, Kong X, Krasieva TB, Ball AR Jr, Yokomori K. Kim JS, et al. Methods Cell Biol. 2007;82:377-407. doi: 10.1016/S0091-679X(06)82013-3. Methods Cell Biol. 2007. PMID: 17586265 - Application of Laser Micro-irradiation for Examination of Single and Double Strand Break Repair in Mammalian Cells.
Holton NW, Andrews JF, Gassman NR. Holton NW, et al. J Vis Exp. 2017 Sep 5;(127):56265. doi: 10.3791/56265. J Vis Exp. 2017. PMID: 28930988 Free PMC article. - Somatic cell mutations caused by 365 nm LED-UVA due to DNA double-strand breaks through oxidative damage.
Fang X, Ide N, Higashi S, Kamei Y, Toyooka T, Ibuki Y, Kawai K, Kasai H, Okamoto K, Arimoto-Kobayashi S, Negishi T. Fang X, et al. Photochem Photobiol Sci. 2014 Sep;13(9):1338-46. doi: 10.1039/c4pp00148f. Photochem Photobiol Sci. 2014. PMID: 25027494 - Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage - when is a DSB not a DSB?
Reynolds P, Botchway SW, Parker AW, O'Neill P. Reynolds P, et al. Mutat Res. 2013 Aug 30;756(1-2):14-20. doi: 10.1016/j.mrgentox.2013.05.006. Epub 2013 May 17. Mutat Res. 2013. PMID: 23688615 Free PMC article. Review.
Cited by
- The function of BCL11B in base excision repair contributes to its dual role as an oncogene and a haplo-insufficient tumor suppressor gene.
Vickridge E, Faraco CCF, Lo F, Rahimian H, Liu ZY, Tehrani PS, Djerir B, Ramdzan ZM, Leduy L, Maréchal A, Gingras AC, Nepveu A. Vickridge E, et al. Nucleic Acids Res. 2024 Jan 11;52(1):223-242. doi: 10.1093/nar/gkad1037. Nucleic Acids Res. 2024. PMID: 37956270 Free PMC article. - Damage site chromatin: open or closed?
Ball AR Jr, Yokomori K. Ball AR Jr, et al. Curr Opin Cell Biol. 2011 Jun;23(3):277-83. doi: 10.1016/j.ceb.2011.03.012. Epub 2011 Apr 12. Curr Opin Cell Biol. 2011. PMID: 21489773 Free PMC article. Review. - Revisiting the role of heterochromatin protein 1 in DNA repair.
Ball AR Jr, Yokomori K. Ball AR Jr, et al. J Cell Biol. 2009 May 18;185(4):573-5. doi: 10.1083/jcb.200904033. J Cell Biol. 2009. PMID: 19451270 Free PMC article. Review. - Discrimination of Kinetic Models by a Combination of Microirradiation and Fluorescence Photobleaching.
Lengert L, Lengert N, Drossel B, Cardoso MC, Muster B, Nowak D, Rapp A. Lengert L, et al. Biophys J. 2015 Oct 20;109(8):1551-64. doi: 10.1016/j.bpj.2015.08.031. Biophys J. 2015. PMID: 26488646 Free PMC article. - Shining Light in Mechanobiology: Optical Tweezers, Scissors, and Beyond.
Stilgoe AB, Favre-Bulle IA, Watson ML, Gomez-Godinez V, Berns MW, Preece D, Rubinsztein-Dunlop H. Stilgoe AB, et al. ACS Photonics. 2024 Mar 11;11(3):917-940. doi: 10.1021/acsphotonics.4c00064. eCollection 2024 Mar 20. ACS Photonics. 2024. PMID: 38523746 Free PMC article. Review.
References
- Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochemie. 2003;85:1101–1111. - PubMed
- Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. - PubMed
- Taylor AM. Chromosome instability syndromes. Best Pract. Res. Clin. Haematol. 2001;2001:631–644. - PubMed
- Mazin AV, Bornarth CJ, Solinger JA, Heyer WD, Kowalczykowski SC. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. - PubMed
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RR01192/RR/NCRR NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- R25 GM055246/GM/NIGMS NIH HHS/United States
- MBRS-GM-55246/GM/NIGMS NIH HHS/United States
- CA100710/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous