dndDB: a database focused on phosphorothioation of the DNA backbone - PubMed (original) (raw)
dndDB: a database focused on phosphorothioation of the DNA backbone
Hong-Yu Ou et al. PLoS One. 2009.
Abstract
Background: The Dnd DNA degradation phenotype was first observed during electrophoresis of genomic DNA from Streptomyces lividans more than 20 years ago. It was subsequently shown to be governed by the five-gene dnd cluster. Similar gene clusters have now been found to be widespread among many other distantly related bacteria. Recently the dnd cluster was shown to mediate the incorporation of sulphur into the DNA backbone via a sequence-selective, stereo-specific phosphorothioate modification in Escherichia coli B7A. Intriguingly, to date all identified dnd clusters lie within mobile genetic elements, the vast majority in laterally transferred genomic islands.
Methodology: We organized available data from experimental and bioinformatics analyses about the DNA phosphorothioation phenomenon and associated documentation as a dndDB database. It contains the following detailed information: (i) Dnd phenotype; (ii) dnd gene clusters; (iii) genomic islands harbouring dnd genes; (iv) Dnd proteins and conserved domains. As of 25 December 2008, dndDB contained data corresponding to 24 bacterial species exhibiting the Dnd phenotype reported in the scientific literature. In addition, via in silico analysis, dndDB identified 26 syntenic dnd clusters from 25 species of Eubacteria and Archaea, 25 dnd-bearing genomic islands and one dnd plasmid containing 114 dnd genes. A further 397 other genes coding for proteins with varying levels of similarity to Dnd proteins were also included in dndDB. A broad range of similarity search, sequence alignment and phylogenetic tools are readily accessible to allow for to individualized directions of research focused on dnd genes.
Conclusion: dndDB can facilitate efficient investigation of a wide range of aspects relating to dnd DNA modification and other island-encoded functions in host organisms. dndDB version 1.0 is freely available at http://mml.sjtu.edu.cn/dndDB/.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
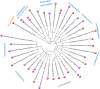
Figure 1. Inferred phylogenetic relationship of the 31 bacterial and one archael organism carrying known dnd clusters (denoted by orange ‘G’ balls) and/or documented to exhibit the Dnd phenotype (denoted by purple ‘P’ balls).
The tree shown was constructed on the basis of NCBI taxonomy (
http://www.ncbi.nlm.nih.gov/Taxonomy/
) by using iTOL , which is now accessible via _dnd_DB.
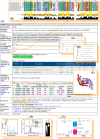
Figure 2. The dnd island in the Salmonella enterica serovar Saintpaul SARA23 genome that is currently being sequenced.
(A) The top axis corresponds to the dnd island-bearing contig (NCBI Refseq accession no. NZ_ABAM01000005), while the lower axis represents a magnified view of the region shown in the red box. The symbol ‘k’ in the coordinates denotes kilobase pairs. (B) A schematic view of the lower axis (above) illustrating the location of the 19.7-kb dnd island (orange line), the leuX tRNA gene integration site (red arrow head), and the upstream/downstream flanking regions (black lines) that are conserved across 14 completely sequenced Salmonella enterica genomes. The ‘5end’ and ‘3end’ backbone labels refer to the 5′- and 3′-flanking backbone segments in relation to the orientation of the leuX tRNA gene, respectively. (C) SynView-facilated synteny mapping of the dnd islands and immediate flanking sequences from three species: Salmonella enterica serovar Saintpaul SARA23 (19.7-kb island) [topmost], Escherichia coli B7A (17.9-kb tRNA-proximal end of island) [middle] and Enterobacter sp. 638 (16.9-kb island) [lower most]. The dnd genes are highlighted in blue, while these and other island-harboured genes are marked by orange frames. Individual genes are hyperlinked to related information that can be accessed using GBrowse. Light-blue-shaded trapezoids link orthologous genes between the three species.
Figure 3. Organization of Dnd proteins and conserved domains in _dnd_DB.
(A) DndA protein data that have been used to predict its putative biological function as a likely cysteine desulfur-transferase in Streptomyces lividans. (B) Multiple amino acid sequence alignment of DndA proteins highlighted the conserved domain in Pfam (accession no. PF00266). (C) and (D) Phylogenetic trees drawn on basis of DndA amino acid sequences and 16S rDNA sequences of the host organisms, respectively. (E) A 3-D structural image corresponding to a DndA-related protein (PDB ID: 1p3w). (G) Sample experimental data demonstrating that DndA provides sulphur via its L-cysteine desulfurylase activity . (F) Inferred biochemical reaction, in which DndA is predicted to catalyze the assembly of DndC as an iron–sulfur cluster protein .
Similar articles
- The dnd operon for DNA phosphorothioation modification system in Escherichia coli is located in diverse genomic islands.
Ho WS, Ou HY, Yeo CC, Thong KL. Ho WS, et al. BMC Genomics. 2015 Mar 17;16(1):199. doi: 10.1186/s12864-015-1421-8. BMC Genomics. 2015. PMID: 25879448 Free PMC article. - DNA phosphorothioation in Streptomyces lividans: mutational analysis of the dnd locus.
Xu T, Liang J, Chen S, Wang L, He X, You D, Wang Z, Li A, Xu Z, Zhou X, Deng Z. Xu T, et al. BMC Microbiol. 2009 Feb 20;9:41. doi: 10.1186/1471-2180-9-41. BMC Microbiol. 2009. PMID: 19232083 Free PMC article. - A novel DNA modification by sulphur.
Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. Zhou X, et al. Mol Microbiol. 2005 Sep;57(5):1428-38. doi: 10.1111/j.1365-2958.2005.04764.x. Mol Microbiol. 2005. PMID: 16102010 - Twenty years hunting for sulfur in DNA.
Chen S, Wang L, Deng Z. Chen S, et al. Protein Cell. 2010 Jan;1(1):14-21. doi: 10.1007/s13238-010-0009-y. Epub 2010 Feb 7. Protein Cell. 2010. PMID: 21203994 Free PMC article. Review. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
- The dnd operon for DNA phosphorothioation modification system in Escherichia coli is located in diverse genomic islands.
Ho WS, Ou HY, Yeo CC, Thong KL. Ho WS, et al. BMC Genomics. 2015 Mar 17;16(1):199. doi: 10.1186/s12864-015-1421-8. BMC Genomics. 2015. PMID: 25879448 Free PMC article. - Phosphorothioate DNA as an antioxidant in bacteria.
Xie X, Liang J, Pu T, Xu F, Yao F, Yang Y, Zhao YL, You D, Zhou X, Deng Z, Wang Z. Xie X, et al. Nucleic Acids Res. 2012 Oct;40(18):9115-24. doi: 10.1093/nar/gks650. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772986 Free PMC article. - Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri.
Van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA. Van Pijkeren JP, et al. Bioengineered. 2012 Jul-Aug;3(4):209-17. doi: 10.4161/bioe.21049. Epub 2012 Jul 1. Bioengineered. 2012. PMID: 22750793 Free PMC article. - Structural basis for the recognition of sulfur in phosphorothioated DNA.
Liu G, Fu W, Zhang Z, He Y, Yu H, Wang Y, Wang X, Zhao YL, Deng Z, Wu G, He X. Liu G, et al. Nat Commun. 2018 Nov 8;9(1):4689. doi: 10.1038/s41467-018-07093-1. Nat Commun. 2018. PMID: 30409991 Free PMC article. - The origin and impeded dissemination of the DNA phosphorothioation system in prokaryotes.
Jian H, Xu G, Yi Y, Hao Y, Wang Y, Xiong L, Wang S, Liu S, Meng C, Wang J, Zhang Y, Chen C, Feng X, Luo H, Zhang H, Zhang X, Wang L, Wang Z, Deng Z, Xiao X. Jian H, et al. Nat Commun. 2021 Nov 4;12(1):6382. doi: 10.1038/s41467-021-26636-7. Nat Commun. 2021. PMID: 34737280 Free PMC article.
References
- Zhou X, He X, Liang J, Li A, Xu T, et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. - PubMed
- Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. - PubMed
- He X, Ou HY, Yu Q, Zhou X, Wu J, et al. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65:1034–1048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials