Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei - PubMed (original) (raw)
Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei
S de Nooijer et al. Nucleic Acids Res. 2009 Jun.
Abstract
The organization of the eukaryote nucleus into functional compartments arises by self-organization both through specific protein-protein and protein-DNA interactions and non-specific interactions that lead to entropic effects, such as e.g. depletion attraction. While many specific interactions have so far been demonstrated, the contributions of non-specific interactions are still unclear. We used coarse-grained molecular dynamics simulations of previously published models for Arabidopsis thaliana chromatin organization to show that non-specific interactions can explain the in vivo localization of nucleoli and chromocenters. Also, we quantitatively demonstrate that chromatin looping contributes to the formation of chromosome territories. Our results are consistent with the previously published Rosette model for Arabidopsis chromatin organization and suggest that chromocenter-associated loops play a role in suppressing chromocenter clustering.
Figures
Figure 1.
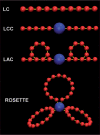
Graphical depiction of the various models. The most simple model is the linear chain (LC) model, in which chromosomes are modelled as consisting of identical monomers (red) arranged in linear chains with harmonic spring potentials (yellow) connecting the monomers. The linear chains with chromocenters (LCC) model is almost identical to the LC model, but models the centromeric area of the chromocome as a large chromocenter (blue). An expansion of the LCC model is the looped arms with chromocenters model (LAC), in which the chains contain loops. In the Rosette model (after Fransz et al. 2002) the chromosome arms loop out from a chromocenter in several loops. Chromocenters, monomers and bonds are not drawn according to scale.
Figure 2.
Overview showing orthographically rendered snapshots of single configurations from LCC (top row), LAC (middle row) and Rosette (bottom row) model simulations containing 10 chromosomes, with chromocenters, in different colors. Of each configuration, three images are provided: left column shows all monomers in the simulation, middle column shows one chromosome, and the right column shows the localization of chromocenters and nucleoli (brown) only.
Figure 3.
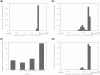
Chromocenter localization and clustering. (a) Histogram showing the fraction of chromocenters in each radial position bin in a LCC model simulation. (b) Distribution of chromocenter radial positions in the Rosette model. (c) Clustering analysis on the simulations of (a and b) showing the average amount of chromocenter clusters in each simulation. (d) Distribution of chromocenter radial positions in the rosette model including a 1.5 μm nucleolus.
Figure 4.
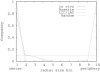
Nucleolus positions in Arabidopsis. On the horizontal axis nucleolus position bins are shown each representing a shell of 0.1 times the nuclear volume available to the nucleolus. On the vertical axis, the fraction of nucleoli in each bin is shown. The solid line shows measured positions in A. thaliana mesophyl nuclei, the dashed line shows the prediction derived from the Rosette model, the line with alternating dashes and dots shows predictions from the LAC model, and the dotted line shows random localization (assuming the nucleolus to have an equal chance to localize to every available position in the nucleus).
Similar articles
- Nuclear organization in crucifer genomes: nucleolus-associated telomere clustering is not a universal interphase configuration in Brassicaceae.
Shan W, Kubová M, Mandáková T, Lysak MA. Shan W, et al. Plant J. 2021 Oct;108(2):528-540. doi: 10.1111/tpj.15459. Epub 2021 Sep 12. Plant J. 2021. PMID: 34390055 - Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate.
Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Fransz P, et al. Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14584-9. doi: 10.1073/pnas.212325299. Epub 2002 Oct 16. Proc Natl Acad Sci U S A. 2002. PMID: 12384572 Free PMC article. - Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness.
Schubert V, Berr A, Meister A. Schubert V, et al. Chromosoma. 2012 Aug;121(4):369-87. doi: 10.1007/s00412-012-0367-8. Epub 2012 Apr 4. Chromosoma. 2012. PMID: 22476443 - Heterochromatin in interphase nuclei of Arabidopsis thaliana.
Fransz P, Soppe W, Schubert I. Fransz P, et al. Chromosome Res. 2003;11(3):227-40. doi: 10.1023/a:1022835825899. Chromosome Res. 2003. PMID: 12769290 Review. - Nuclear architecture and chromatin dynamics in interphase nuclei of Arabidopsis thaliana.
Del Prete S, Arpón J, Sakai K, Andrey P, Gaudin V. Del Prete S, et al. Cytogenet Genome Res. 2014;143(1-3):28-50. doi: 10.1159/000363724. Epub 2014 Jun 28. Cytogenet Genome Res. 2014. PMID: 24992956 Review.
Cited by
- Molecular simulations of cellular processes.
Trovato F, Fumagalli G. Trovato F, et al. Biophys Rev. 2017 Dec;9(6):941-958. doi: 10.1007/s12551-017-0363-6. Epub 2017 Nov 28. Biophys Rev. 2017. PMID: 29185136 Free PMC article. Review. - Expression-dependent folding of interphase chromatin.
Jerabek H, Heermann DW. Jerabek H, et al. PLoS One. 2012;7(5):e37525. doi: 10.1371/journal.pone.0037525. Epub 2012 May 23. PLoS One. 2012. PMID: 22649534 Free PMC article. - Consistencies and contradictions in different polymer models of chromatin architecture.
Câmara AS, Mascher M. Câmara AS, et al. Comput Struct Biotechnol J. 2023 Jan 24;21:1084-1091. doi: 10.1016/j.csbj.2023.01.033. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36789261 Free PMC article. Review. - Investigation of the chromosome regions with significant affinity for the nuclear envelope in fruit fly--a model based approach.
Kinney NA, Sharakhov IV, Onufriev AV. Kinney NA, et al. PLoS One. 2014 Mar 20;9(3):e91943. doi: 10.1371/journal.pone.0091943. eCollection 2014. PLoS One. 2014. PMID: 24651400 Free PMC article. - Perinuclear distribution of heterochromatin in developing C. elegans embryos.
Grant J, Verrill C, Coustham V, Arneodo A, Palladino F, Monier K, Khalil A. Grant J, et al. Chromosome Res. 2010 Dec;18(8):873-85. doi: 10.1007/s10577-010-9175-2. Epub 2010 Nov 30. Chromosome Res. 2010. PMID: 21116703
References
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum. Genet. 1982;62:201–209. - PubMed
- Handwerger K, Gall J. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. - PubMed
- Ye Q, Callebaut I, Pezhman A, Courvalin J, Worman H. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997;272:14983–14989. - PubMed